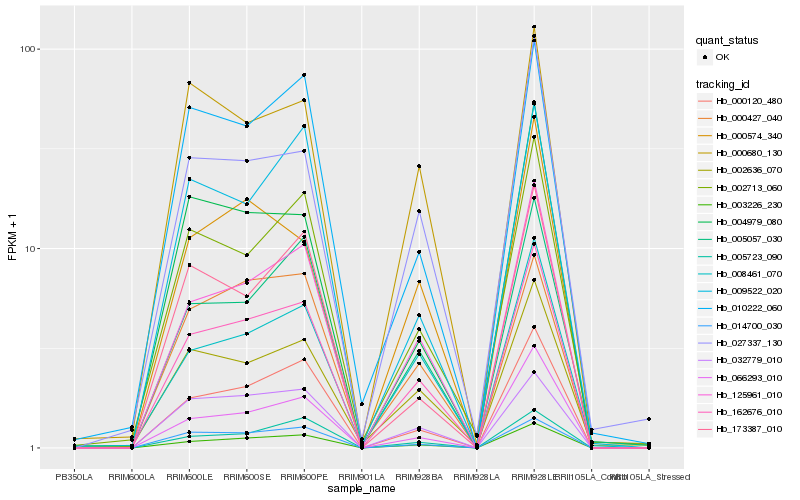
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162676_010 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_003226_230 |
0.0338066371 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 3 |
Hb_125961_010 |
0.0509812578 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000120_480 |
0.0666924027 |
- |
- |
- |
| 5 |
Hb_005057_030 |
0.0892809784 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_014700_030 |
0.0927565758 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 7 |
Hb_008461_070 |
0.0930417671 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002713_060 |
0.0971907326 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 9 |
Hb_066293_010 |
0.0973157569 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_005723_090 |
0.0998301345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002636_070 |
0.1016240459 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 12 |
Hb_032779_010 |
0.1081502773 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_010222_060 |
0.1097163647 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_027337_130 |
0.1116858842 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 15 |
Hb_000680_130 |
0.1125099681 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_000574_340 |
0.1158429455 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 17 |
Hb_173387_010 |
0.1213325783 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 18 |
Hb_000427_040 |
0.1234703567 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 19 |
Hb_004979_080 |
0.1235102037 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_009522_020 |
0.1235739612 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |