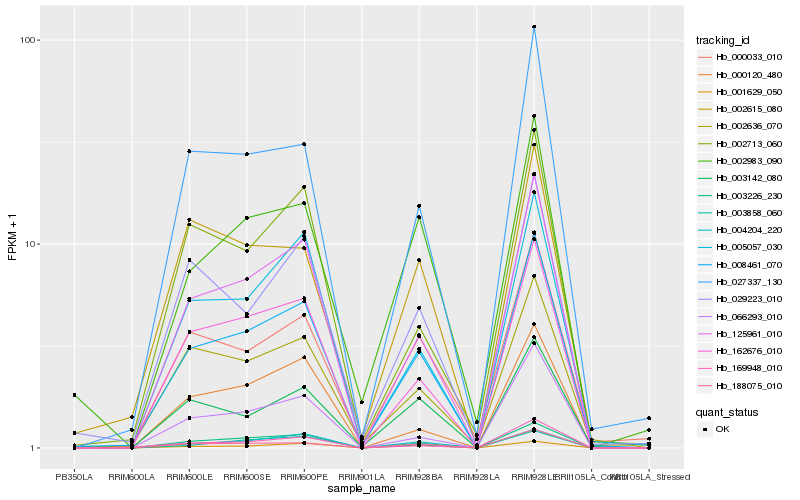
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008461_070 |
0.0 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_125961_010 |
0.0723261445 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_188075_010 |
0.0908902486 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 4 |
Hb_162676_010 |
0.0930417671 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_027337_130 |
0.0974033754 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 6 |
Hb_003226_230 |
0.0986307972 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 7 |
Hb_002636_070 |
0.0995220305 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 8 |
Hb_005057_030 |
0.1040774158 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_000033_010 |
0.1068649543 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 10 |
Hb_003142_080 |
0.1088772235 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_029223_010 |
0.1129489706 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 12 |
Hb_004204_220 |
0.1185151188 |
- |
- |
- |
| 13 |
Hb_066293_010 |
0.123127401 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 14 |
Hb_003858_060 |
0.1250881934 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 15 |
Hb_002713_060 |
0.1252165771 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 16 |
Hb_002983_090 |
0.1257798895 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 17 |
Hb_001629_050 |
0.1293105997 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 18 |
Hb_000120_480 |
0.1300768274 |
- |
- |
- |
| 19 |
Hb_169948_010 |
0.1310176272 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 20 |
Hb_002615_080 |
0.1328835166 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |