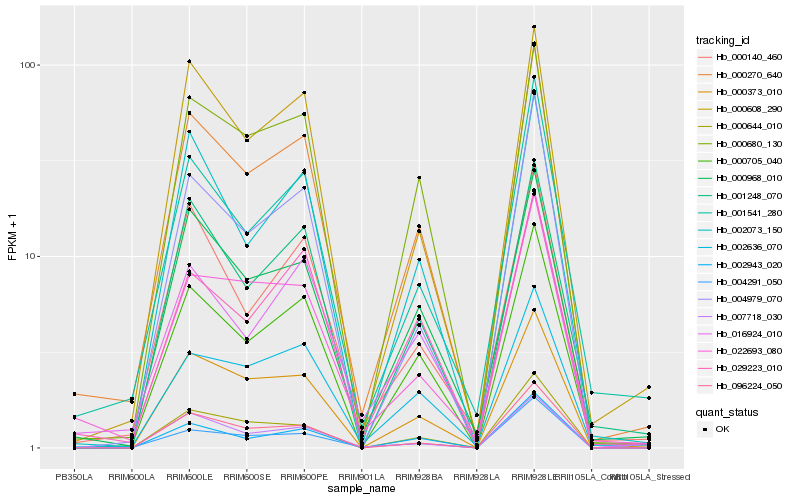
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_640 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 2 |
Hb_000705_040 |
0.0736966471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001541_280 |
0.0821011605 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 4 |
Hb_000968_010 |
0.0842916181 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000373_010 |
0.0872407619 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_002636_070 |
0.0902684821 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 7 |
Hb_004979_070 |
0.0923235869 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 8 |
Hb_007718_030 |
0.0942335909 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000680_130 |
0.0943889889 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 10 |
Hb_000608_290 |
0.0950500361 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 11 |
Hb_016924_010 |
0.0975739736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002943_020 |
0.0982228918 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 13 |
Hb_096224_050 |
0.09833366 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 14 |
Hb_004291_050 |
0.0992276749 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 15 |
Hb_000140_460 |
0.0992730112 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 16 |
Hb_029223_010 |
0.0995542048 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 17 |
Hb_002073_150 |
0.099577761 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 18 |
Hb_022693_080 |
0.1011889158 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 19 |
Hb_000644_010 |
0.1013395464 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001248_070 |
0.1020607387 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |