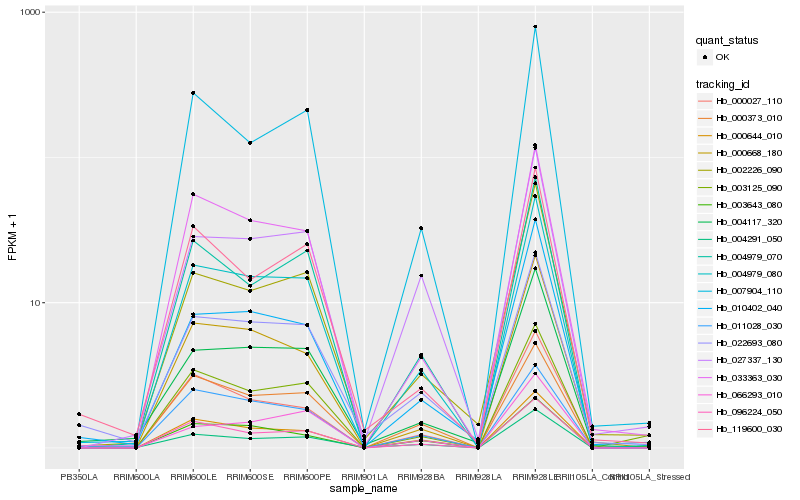
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004979_080 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_096224_050 |
0.0644435594 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 3 |
Hb_000644_010 |
0.0732765834 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004979_070 |
0.0753218105 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 5 |
Hb_033363_030 |
0.075932204 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007904_110 |
0.0769514918 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_000668_180 |
0.0853619746 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 8 |
Hb_004291_050 |
0.0854569129 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 9 |
Hb_000027_110 |
0.092820048 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 10 |
Hb_003643_080 |
0.0953101028 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 11 |
Hb_004117_320 |
0.0975939377 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 12 |
Hb_000373_010 |
0.0988694625 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_002226_090 |
0.0993642784 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 14 |
Hb_011028_030 |
0.1008618729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_022693_080 |
0.1042572769 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 16 |
Hb_010402_040 |
0.1044400269 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 17 |
Hb_003125_090 |
0.1084332531 |
- |
- |
- |
| 18 |
Hb_066293_010 |
0.1107186993 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 19 |
Hb_027337_130 |
0.1111793076 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 20 |
Hb_119600_030 |
0.1119464319 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |