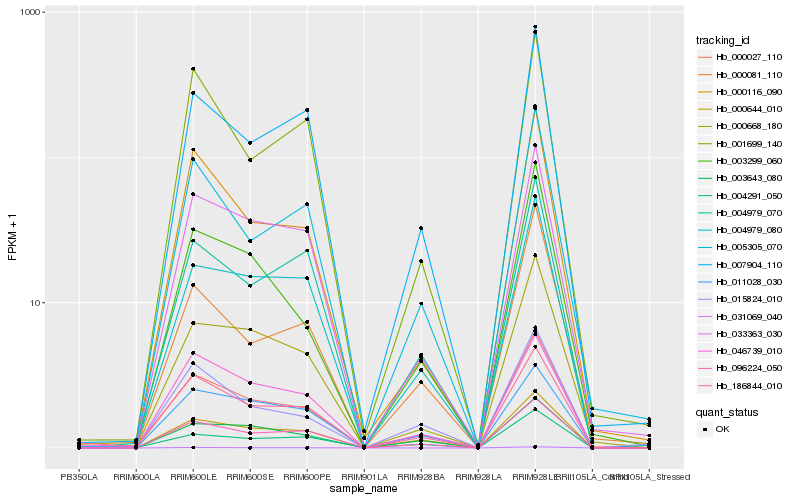
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_110 |
0.0 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 2 |
Hb_096224_050 |
0.0684354474 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 3 |
Hb_000644_010 |
0.0748396811 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_033363_030 |
0.0775308851 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000668_180 |
0.0814272471 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 6 |
Hb_000116_090 |
0.0889629148 |
- |
- |
catalase [Hevea brasiliensis] |
| 7 |
Hb_005305_070 |
0.0898070325 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 8 |
Hb_007904_110 |
0.0927098866 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_004979_080 |
0.092820048 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_003299_060 |
0.0993448212 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_003643_080 |
0.1008205432 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 12 |
Hb_186844_010 |
0.1034981172 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_015824_010 |
0.1041733098 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_004979_070 |
0.1077639448 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 15 |
Hb_001699_140 |
0.1113710714 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 16 |
Hb_046739_010 |
0.1117002094 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 17 |
Hb_031069_040 |
0.1144358351 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000081_110 |
0.1149377212 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_004291_050 |
0.1154187876 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 20 |
Hb_011028_030 |
0.1176098173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |