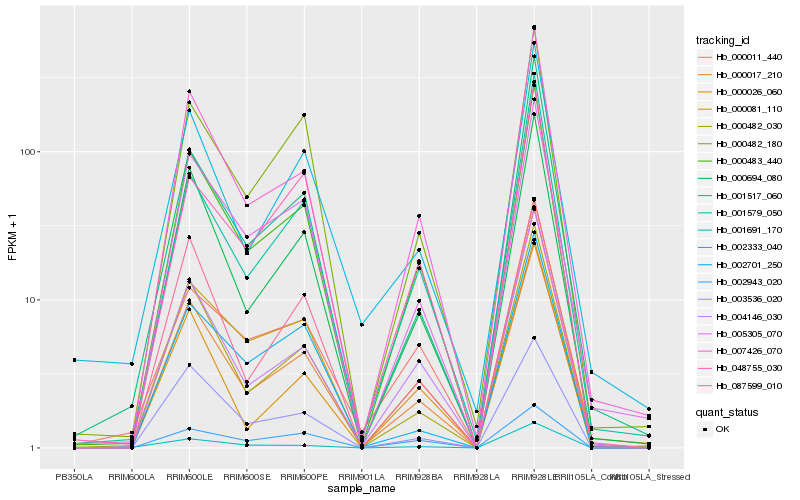
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_440 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000017_210 |
0.0408143377 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 3 |
Hb_007426_070 |
0.0543005576 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 4 |
Hb_000694_080 |
0.0680858125 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 5 |
Hb_000081_110 |
0.0762542943 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_004146_030 |
0.0774857721 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_005305_070 |
0.0809405042 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 8 |
Hb_000482_030 |
0.082401989 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000482_180 |
0.089520379 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 10 |
Hb_003536_020 |
0.0994659145 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 11 |
Hb_001579_050 |
0.099867474 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000011_440 |
0.1016923708 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 13 |
Hb_087599_010 |
0.1051338788 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 14 |
Hb_048755_030 |
0.1073091889 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_001691_170 |
0.1082022157 |
- |
- |
PREDICTED: callose synthase 12-like [Glycine max] |
| 16 |
Hb_001517_060 |
0.1082820269 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 17 |
Hb_002333_040 |
0.1102019977 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 18 |
Hb_002943_020 |
0.1114796034 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 19 |
Hb_002701_250 |
0.1116997068 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_000026_060 |
0.1133331703 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |