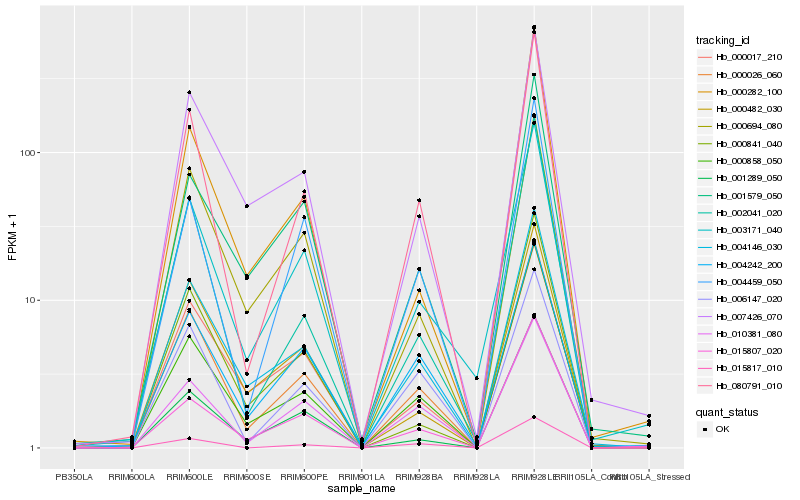
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000026_060 |
0.0 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |
| 2 |
Hb_080791_010 |
0.0449856245 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 3 |
Hb_004146_030 |
0.0621521593 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000858_050 |
0.0845620042 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_003171_040 |
0.0891741278 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 6 |
Hb_015817_010 |
0.0955606625 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 7 |
Hb_007426_070 |
0.0965227487 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 8 |
Hb_006147_020 |
0.0993865336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002041_020 |
0.099721566 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 10 |
Hb_000841_040 |
0.1029411919 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000694_080 |
0.1047597999 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 12 |
Hb_001289_050 |
0.1049568936 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 13 |
Hb_004459_050 |
0.1069525657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004242_200 |
0.1071213416 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 15 |
Hb_000482_030 |
0.107962388 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000017_210 |
0.1086599915 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 17 |
Hb_001579_050 |
0.110423879 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_015807_020 |
0.1107856915 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 19 |
Hb_010381_080 |
0.1122193578 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000282_100 |
0.1125844307 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |