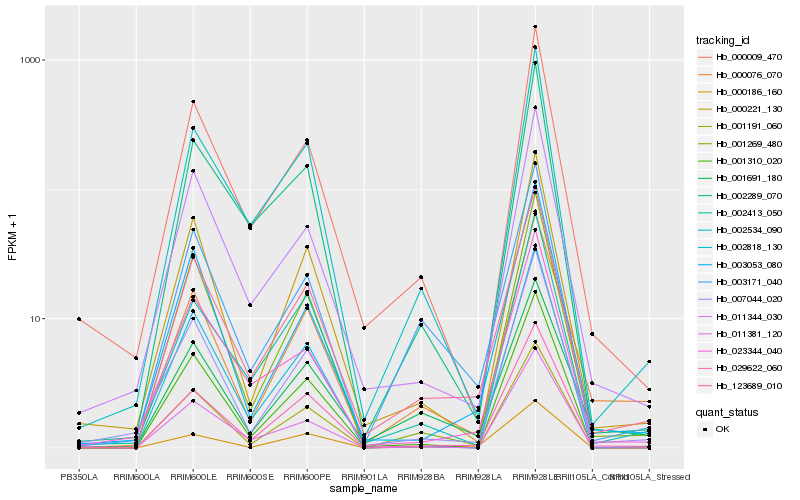
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123689_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 2 |
Hb_029622_060 |
0.0731348138 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002534_090 |
0.0847381511 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 4 |
Hb_000009_470 |
0.0915048779 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_011344_030 |
0.0917880438 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007044_020 |
0.0924775276 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001269_480 |
0.0930588244 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000221_130 |
0.0947130907 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003171_040 |
0.0953918311 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_001691_180 |
0.0964766976 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 11 |
Hb_001310_020 |
0.0987648772 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000076_070 |
0.0989141063 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 13 |
Hb_002289_070 |
0.0998493851 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 14 |
Hb_023344_040 |
0.1006743957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003053_080 |
0.102064345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002413_050 |
0.1030080643 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 17 |
Hb_001191_060 |
0.1072395812 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 18 |
Hb_011381_120 |
0.1072464515 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 19 |
Hb_000186_160 |
0.1076781212 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 20 |
Hb_002818_130 |
0.1078769687 |
- |
- |
cytochrome P450, putative [Ricinus communis] |