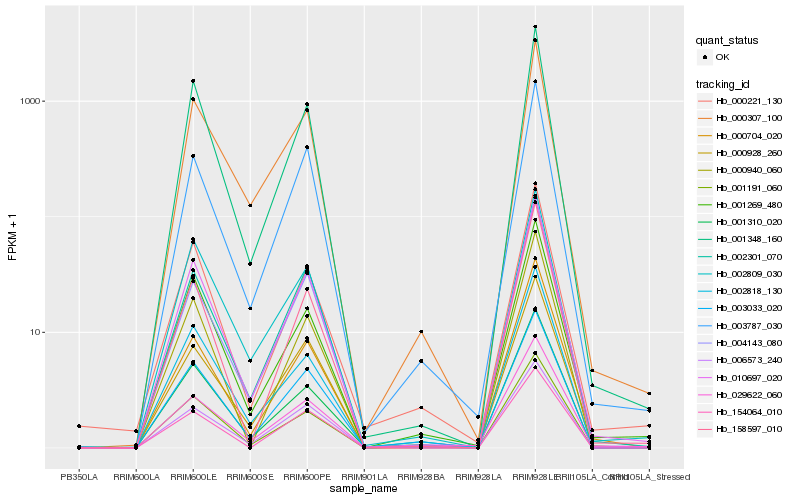
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010697_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001348_160 |
0.0400734588 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_002301_070 |
0.0444646581 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_003787_030 |
0.0550350805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000928_260 |
0.058099263 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 6 |
Hb_001191_060 |
0.0585445153 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 7 |
Hb_001269_480 |
0.0634778365 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000307_100 |
0.0640775627 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003033_020 |
0.064197066 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 10 |
Hb_002809_030 |
0.0646871042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002818_130 |
0.0688426458 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_001310_020 |
0.0694960399 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000704_020 |
0.074066508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006573_240 |
0.0752018481 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_000221_130 |
0.0759799105 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_158597_010 |
0.0759978696 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_004143_080 |
0.0774121398 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 18 |
Hb_000940_060 |
0.077997531 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 19 |
Hb_154064_010 |
0.0788181905 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 20 |
Hb_029622_060 |
0.0790354761 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |