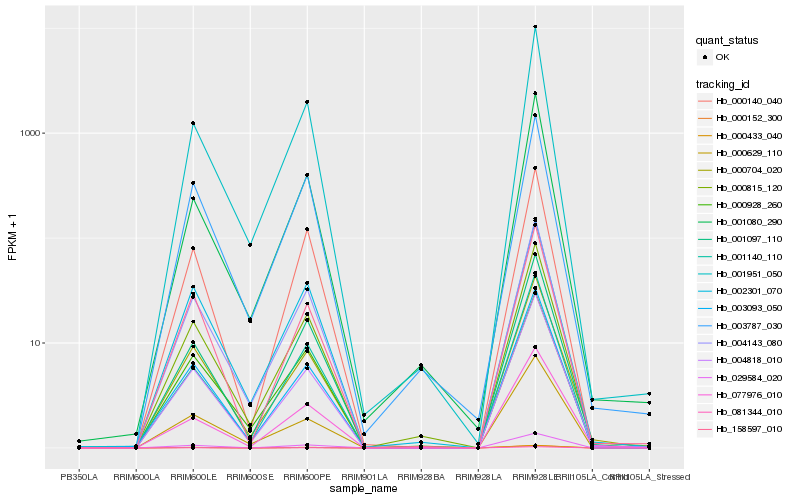
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_120 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_004143_080 |
0.0403622412 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 3 |
Hb_004818_010 |
0.0501754509 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 4 |
Hb_000704_020 |
0.0538655907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001951_050 |
0.0548087561 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 6 |
Hb_002301_070 |
0.0560163602 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_003093_050 |
0.0586099576 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003787_030 |
0.0586933773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001097_110 |
0.0590720427 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 10 |
Hb_000928_260 |
0.0655885375 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 11 |
Hb_077976_010 |
0.0667917169 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 12 |
Hb_000140_040 |
0.0692127637 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 13 |
Hb_158597_010 |
0.0699170532 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_029584_020 |
0.0704292257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_081344_010 |
0.0707579569 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 16 |
Hb_000152_300 |
0.0714972571 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000433_040 |
0.0716916237 |
- |
- |
- |
| 18 |
Hb_001080_290 |
0.0725355749 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 19 |
Hb_000629_110 |
0.0726033904 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 20 |
Hb_001140_110 |
0.0728654723 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |