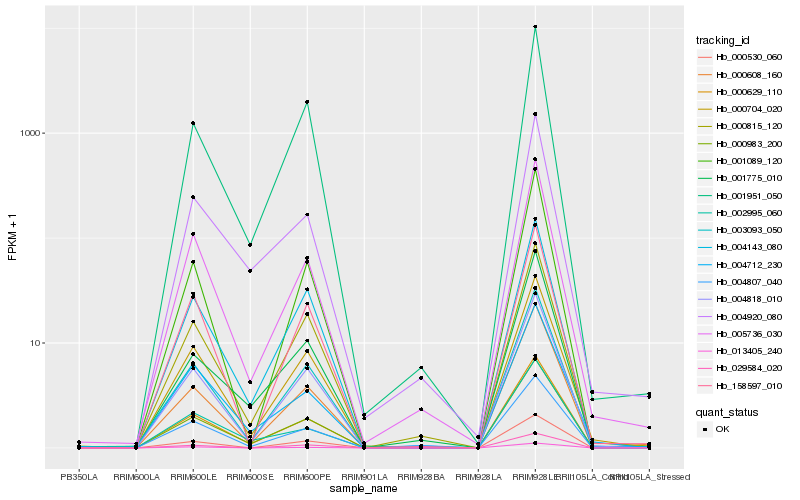
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000629_110 |
0.0 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 2 |
Hb_003093_050 |
0.048247846 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004818_010 |
0.0506913775 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 4 |
Hb_004143_080 |
0.0604933745 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 5 |
Hb_000704_020 |
0.0607033584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000983_200 |
0.0613355713 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 7 |
Hb_005736_030 |
0.065298246 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 8 |
Hb_000608_160 |
0.0653057059 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001951_050 |
0.071676099 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 10 |
Hb_002995_060 |
0.0719361796 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 11 |
Hb_000815_120 |
0.0726033904 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_004920_080 |
0.0730714639 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000530_060 |
0.0756554797 |
- |
- |
- |
| 14 |
Hb_001089_120 |
0.0777370877 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 15 |
Hb_029584_020 |
0.0803149956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004807_040 |
0.0819587371 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 17 |
Hb_158597_010 |
0.0829965041 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001775_010 |
0.0835381435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004712_230 |
0.0853323739 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 20 |
Hb_013405_240 |
0.0860777496 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |