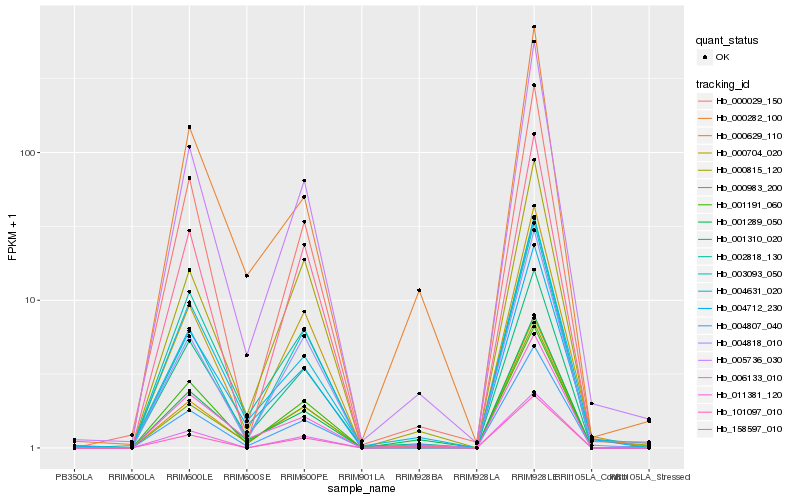
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004807_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 2 |
Hb_001289_050 |
0.0472288319 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 3 |
Hb_101097_010 |
0.0599284096 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_005736_030 |
0.0669721903 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 5 |
Hb_006133_010 |
0.0717123579 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_001310_020 |
0.0734308573 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000029_150 |
0.0780875854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000815_120 |
0.080012686 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000629_110 |
0.0819587371 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 10 |
Hb_000983_200 |
0.0843224031 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 11 |
Hb_003093_050 |
0.0856520936 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001191_060 |
0.0871298459 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 13 |
Hb_011381_120 |
0.0873010268 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 14 |
Hb_004818_010 |
0.0879415006 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 15 |
Hb_158597_010 |
0.0885598953 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000704_020 |
0.0887098289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004712_230 |
0.0902325345 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 18 |
Hb_004631_020 |
0.0912110859 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_000282_100 |
0.0914110011 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 20 |
Hb_002818_130 |
0.0918125701 |
- |
- |
cytochrome P450, putative [Ricinus communis] |