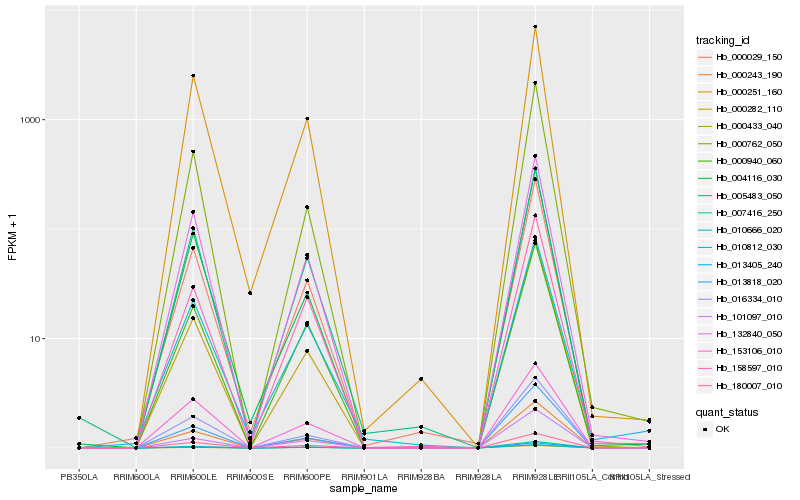
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_190 |
0.0 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 2 |
Hb_013405_240 |
0.0347916922 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 3 |
Hb_007416_250 |
0.0383163798 |
- |
- |
- |
| 4 |
Hb_132840_050 |
0.0386837187 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000940_060 |
0.041257286 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 6 |
Hb_016334_010 |
0.0444765729 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 7 |
Hb_000029_150 |
0.0453851494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005483_050 |
0.0499439026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_180007_010 |
0.0536100255 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_013818_020 |
0.056596566 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 11 |
Hb_158597_010 |
0.0570873084 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_153106_010 |
0.0612762545 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 13 |
Hb_000282_110 |
0.0642185541 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 14 |
Hb_000762_050 |
0.06558784 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000251_160 |
0.0688127077 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 16 |
Hb_004116_030 |
0.0718830851 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 17 |
Hb_010812_030 |
0.0738434866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010666_020 |
0.0755493744 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 19 |
Hb_000433_040 |
0.0756762363 |
- |
- |
- |
| 20 |
Hb_101097_010 |
0.076511414 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |