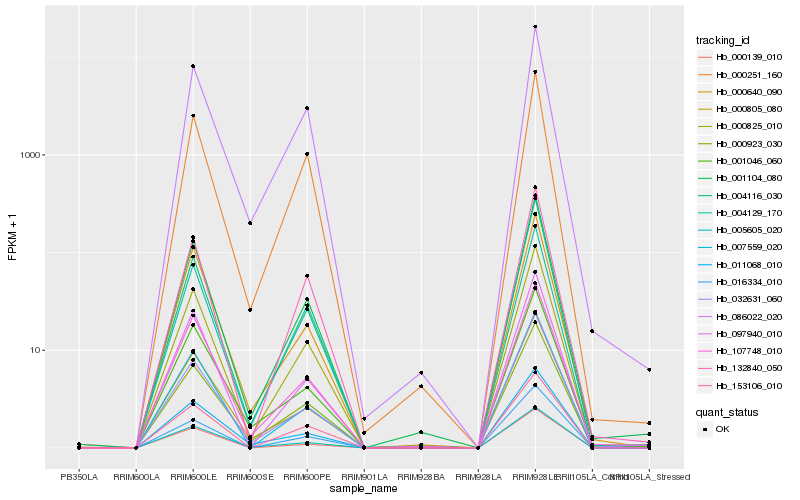
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000825_010 |
0.0 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 2 |
Hb_001104_080 |
0.0291757263 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_000923_030 |
0.0461998289 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 4 |
Hb_000251_160 |
0.0481197514 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 5 |
Hb_007559_020 |
0.0486917619 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 6 |
Hb_132840_050 |
0.0504176592 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005605_020 |
0.0528916702 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 8 |
Hb_153106_010 |
0.0533430296 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 9 |
Hb_004129_170 |
0.0540366116 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 10 |
Hb_016334_010 |
0.0546450538 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 11 |
Hb_097940_010 |
0.0548920833 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 12 |
Hb_000805_080 |
0.0595318182 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_032631_060 |
0.0602057757 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 14 |
Hb_107748_010 |
0.0604457921 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_086022_020 |
0.0605257892 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 16 |
Hb_011068_010 |
0.0621735575 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 17 |
Hb_000139_010 |
0.0622649066 |
- |
- |
- |
| 18 |
Hb_000640_090 |
0.0638019037 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_004116_030 |
0.0646144937 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 20 |
Hb_001046_060 |
0.0652710742 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |