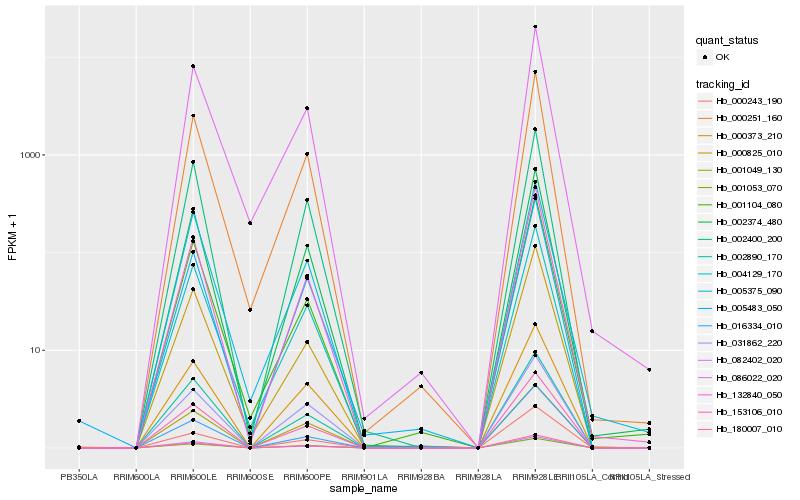
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153106_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 2 |
Hb_180007_010 |
0.0177286791 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_002374_480 |
0.0332346362 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_004129_170 |
0.0373541998 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 5 |
Hb_132840_050 |
0.0381239917 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000251_160 |
0.0382826279 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 7 |
Hb_002890_170 |
0.0512179932 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 8 |
Hb_000373_210 |
0.0520486136 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 9 |
Hb_000825_010 |
0.0533430296 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 10 |
Hb_002400_200 |
0.0543477203 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 11 |
Hb_086022_020 |
0.0610795322 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 12 |
Hb_000243_190 |
0.0612762545 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 13 |
Hb_016334_010 |
0.0636983261 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 14 |
Hb_082402_020 |
0.0641207065 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 15 |
Hb_001053_070 |
0.0641537699 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein EUGRSUZ_B02976 [Eucalyptus grandis] |
| 16 |
Hb_005483_050 |
0.064661364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_031862_220 |
0.0688504951 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 18 |
Hb_001049_130 |
0.0694931778 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_005375_090 |
0.0697655378 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001104_080 |
0.0707200281 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |