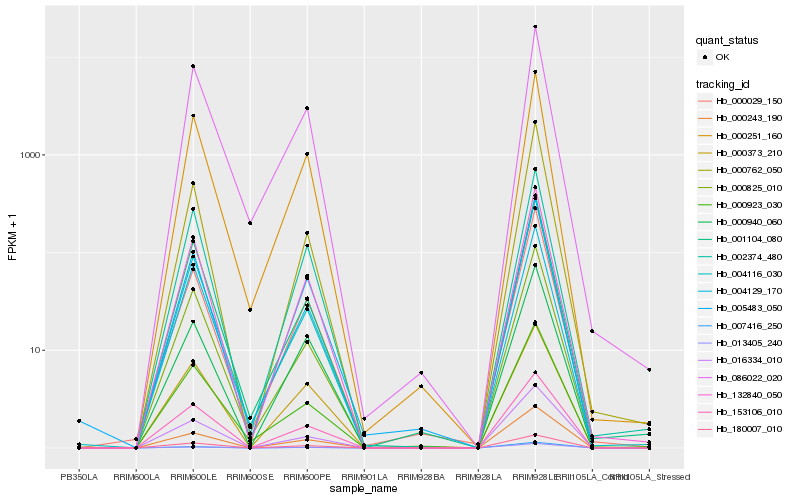
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_050 |
0.0 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 2 |
Hb_153106_010 |
0.0381239917 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 3 |
Hb_180007_010 |
0.0385498756 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_000243_190 |
0.0386837187 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 5 |
Hb_016334_010 |
0.0417105117 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 6 |
Hb_000251_160 |
0.0451927806 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 7 |
Hb_000825_010 |
0.0504176592 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 8 |
Hb_002374_480 |
0.0522459804 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_005483_050 |
0.0551574912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001104_080 |
0.0575098021 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 11 |
Hb_000029_150 |
0.0576145942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004129_170 |
0.0588931449 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 13 |
Hb_000940_060 |
0.0637422728 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 14 |
Hb_013405_240 |
0.0650215261 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 15 |
Hb_000923_030 |
0.065059322 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 16 |
Hb_007416_250 |
0.0673653295 |
- |
- |
- |
| 17 |
Hb_000762_050 |
0.0675656224 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004116_030 |
0.0677127678 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 19 |
Hb_086022_020 |
0.069731527 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 20 |
Hb_000373_210 |
0.0715475429 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |