| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
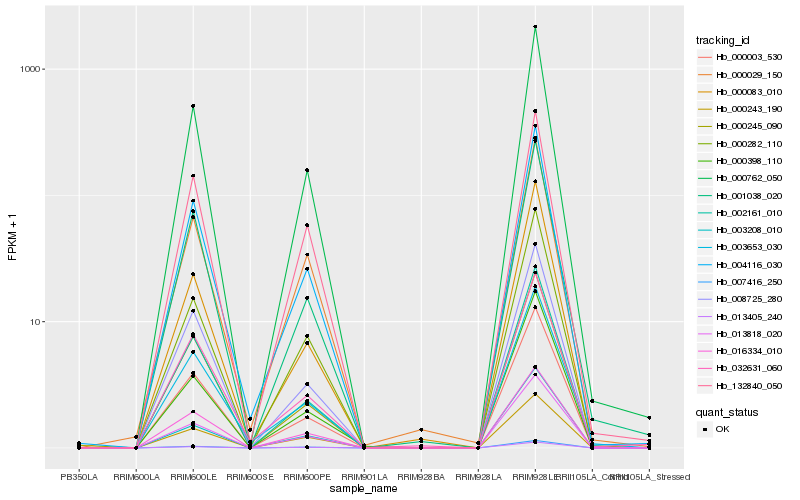
Hb_000762_050 |
0.0 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000003_530 |
0.0251658585 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 3 |
Hb_004116_030 |
0.0320861883 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 4 |
Hb_013818_020 |
0.0334890877 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 5 |
Hb_016334_010 |
0.0379589249 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 6 |
Hb_002161_010 |
0.0385337814 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 7 |
Hb_000282_110 |
0.0443165067 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 8 |
Hb_008725_280 |
0.0444522892 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 9 |
Hb_001038_020 |
0.0466270729 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 10 |
Hb_007416_250 |
0.04939457 |
- |
- |
- |
| 11 |
Hb_013405_240 |
0.0515749133 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 12 |
Hb_000245_090 |
0.0561955758 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 13 |
Hb_003653_030 |
0.0589262169 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000029_150 |
0.0638993551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000398_110 |
0.0655094712 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 16 |
Hb_000243_190 |
0.06558784 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 17 |
Hb_000083_010 |
0.0665609023 |
- |
- |
- |
| 18 |
Hb_003208_010 |
0.0672596739 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_132840_050 |
0.0675656224 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_032631_060 |
0.0692725502 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |