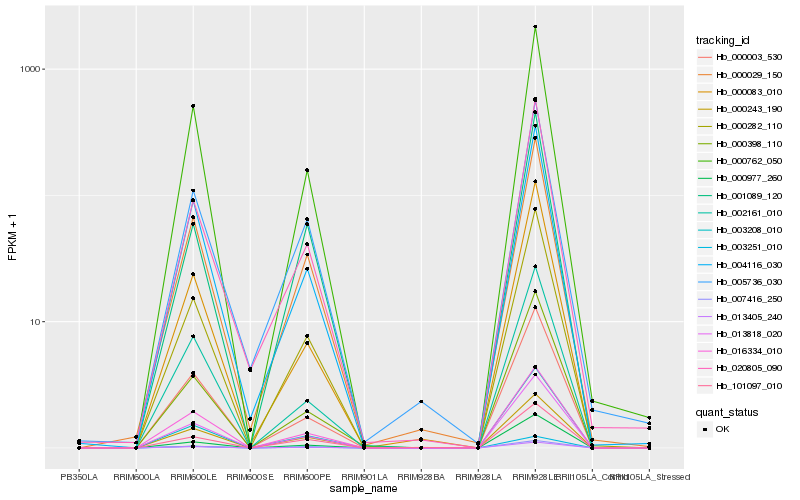
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000282_110 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 2 |
Hb_013818_020 |
0.0214449479 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 3 |
Hb_007416_250 |
0.0287729097 |
- |
- |
- |
| 4 |
Hb_013405_240 |
0.03224312 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 5 |
Hb_003208_010 |
0.0332240383 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_000762_050 |
0.0443165067 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_003251_010 |
0.0478594787 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 8 |
Hb_000398_110 |
0.0520435867 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 9 |
Hb_000977_260 |
0.0538200596 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 10 |
Hb_000003_530 |
0.058128309 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 11 |
Hb_000029_150 |
0.0594707699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_020805_090 |
0.0624687519 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_004116_030 |
0.0629491673 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 14 |
Hb_000243_190 |
0.0642185541 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 15 |
Hb_016334_010 |
0.0644396123 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 16 |
Hb_000083_010 |
0.0680828412 |
- |
- |
- |
| 17 |
Hb_005736_030 |
0.0693716497 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 18 |
Hb_002161_010 |
0.0733115536 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 19 |
Hb_001089_120 |
0.0741108897 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 20 |
Hb_101097_010 |
0.0749824655 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |