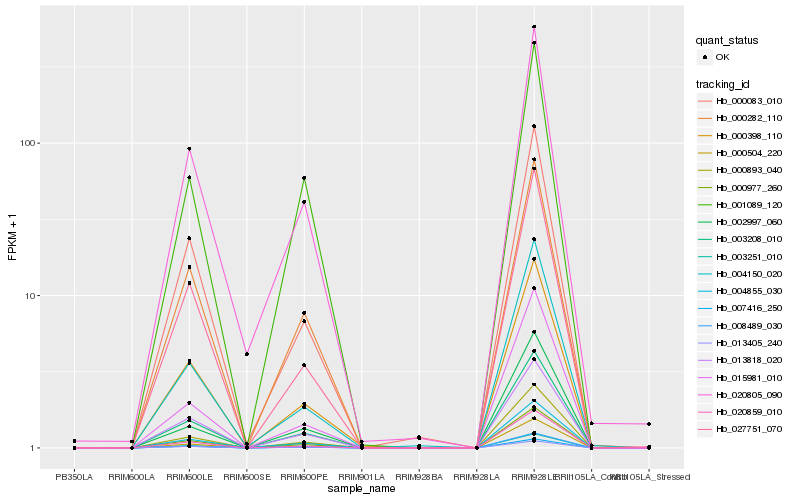
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003251_010 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 2 |
Hb_003208_010 |
0.0186427422 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_000977_260 |
0.0232154197 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 4 |
Hb_000893_040 |
0.0369344737 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 5 |
Hb_000282_110 |
0.0478594787 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 6 |
Hb_008489_030 |
0.0483500796 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 7 |
Hb_000398_110 |
0.0497296165 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 8 |
Hb_001089_120 |
0.0592379157 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 9 |
Hb_013818_020 |
0.0596522528 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 10 |
Hb_020805_090 |
0.0608606724 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_004855_030 |
0.0617027617 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_002997_060 |
0.0620071749 |
- |
- |
hypothetical protein POPTR_0011s10730g [Populus trichocarpa] |
| 13 |
Hb_020859_010 |
0.0651904422 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 14 |
Hb_007416_250 |
0.0665841304 |
- |
- |
- |
| 15 |
Hb_015981_010 |
0.0679930443 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_013405_240 |
0.0698030927 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 17 |
Hb_000083_010 |
0.0709240997 |
- |
- |
- |
| 18 |
Hb_027751_070 |
0.07299625 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000504_220 |
0.0756165158 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 20 |
Hb_004150_020 |
0.0756694768 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |