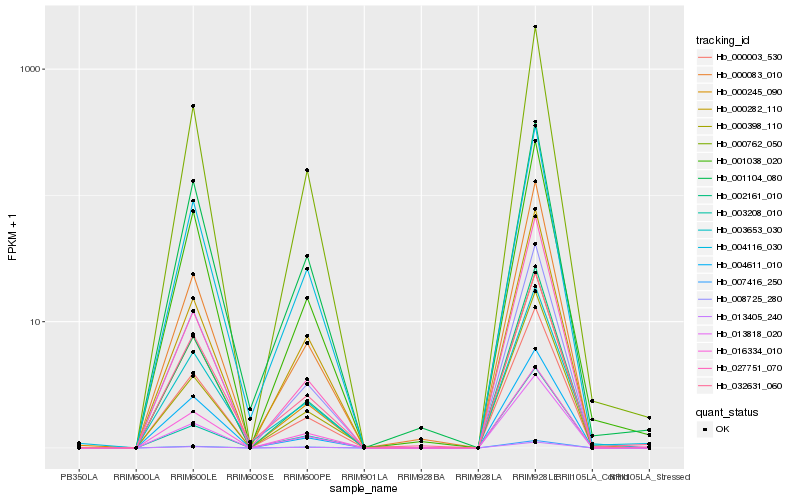
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_530 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 2 |
Hb_002161_010 |
0.0172614332 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 3 |
Hb_000762_050 |
0.0251658585 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_008725_280 |
0.026260106 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 5 |
Hb_004116_030 |
0.03408297 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 6 |
Hb_016334_010 |
0.0401128704 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 7 |
Hb_000245_090 |
0.0414763754 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 8 |
Hb_013818_020 |
0.0417951931 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 9 |
Hb_001038_020 |
0.0461668547 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 10 |
Hb_004611_010 |
0.0555276638 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 11 |
Hb_000282_110 |
0.058128309 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 12 |
Hb_000083_010 |
0.060907362 |
- |
- |
- |
| 13 |
Hb_007416_250 |
0.0614066383 |
- |
- |
- |
| 14 |
Hb_013405_240 |
0.0638041278 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 15 |
Hb_003653_030 |
0.0645463023 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000398_110 |
0.0657300541 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 17 |
Hb_027751_070 |
0.0713855004 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_032631_060 |
0.0718692686 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 19 |
Hb_003208_010 |
0.0736748036 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_001104_080 |
0.0742951007 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |