| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
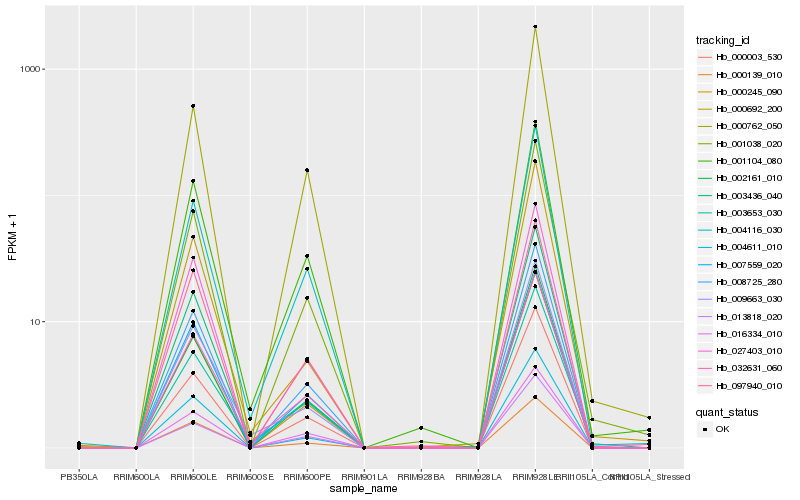
Hb_001038_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 2 |
Hb_008725_280 |
0.0386890715 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 3 |
Hb_002161_010 |
0.0412312961 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 4 |
Hb_000003_530 |
0.0461668547 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 5 |
Hb_000762_050 |
0.0466270729 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000245_090 |
0.0479280599 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 7 |
Hb_004611_010 |
0.0493183198 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 8 |
Hb_004116_030 |
0.0524889238 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 9 |
Hb_032631_060 |
0.0574264124 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_003653_030 |
0.0605289726 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_016334_010 |
0.0625761989 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 12 |
Hb_001104_080 |
0.063275388 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 13 |
Hb_027403_010 |
0.0666134461 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 14 |
Hb_007559_020 |
0.0683116847 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 15 |
Hb_003436_040 |
0.0686679854 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 16 |
Hb_000692_200 |
0.072392028 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 17 |
Hb_097940_010 |
0.0748683151 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 18 |
Hb_009663_030 |
0.075161844 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 19 |
Hb_013818_020 |
0.0770194849 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 20 |
Hb_000139_010 |
0.0787859534 |
- |
- |
- |