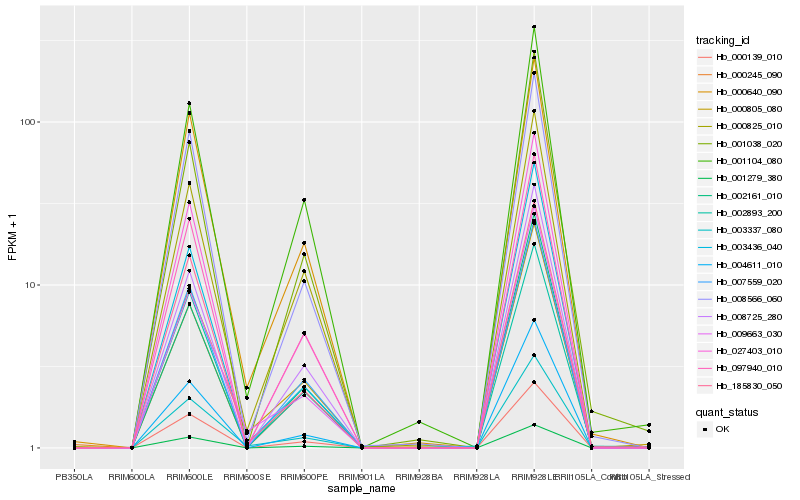
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027403_010 |
0.0 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 2 |
Hb_000139_010 |
0.0251322576 |
- |
- |
- |
| 3 |
Hb_097940_010 |
0.0282060188 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 4 |
Hb_007559_020 |
0.0308592824 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 5 |
Hb_001279_380 |
0.0350584861 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 6 |
Hb_004611_010 |
0.03529574 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_008566_060 |
0.0398553864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_185830_050 |
0.0502175941 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 9 |
Hb_008725_280 |
0.0527195771 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 10 |
Hb_000245_090 |
0.0565203767 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 11 |
Hb_003337_080 |
0.0605418187 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 12 |
Hb_003436_040 |
0.0624368988 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 13 |
Hb_002893_200 |
0.0659327544 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000640_090 |
0.0661085108 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001038_020 |
0.0666134461 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 16 |
Hb_002161_010 |
0.0680968179 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 17 |
Hb_001104_080 |
0.0684541491 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 18 |
Hb_000825_010 |
0.0711983263 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 19 |
Hb_000805_080 |
0.0725715598 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_009663_030 |
0.0748334496 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |