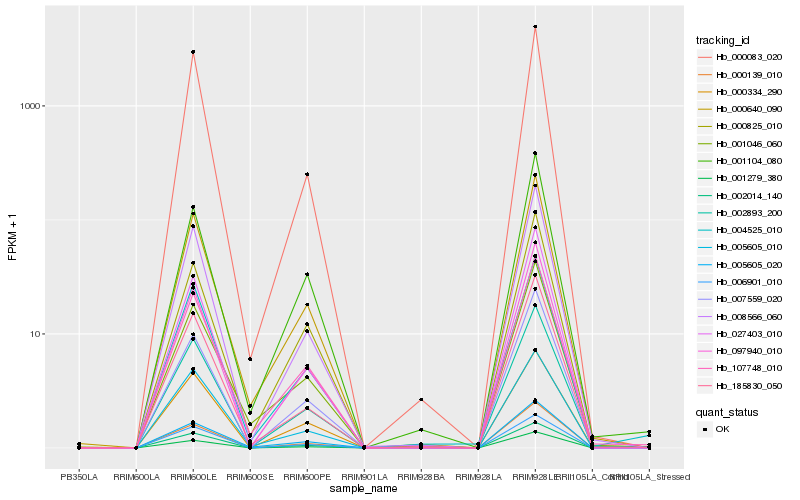
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002893_200 |
0.0 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002014_140 |
0.038487835 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_001279_380 |
0.041078484 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 4 |
Hb_008566_060 |
0.0415786387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000640_090 |
0.0416711519 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_107748_010 |
0.0448655307 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_006901_010 |
0.0465916929 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 8 |
Hb_000139_010 |
0.0486004358 |
- |
- |
- |
| 9 |
Hb_097940_010 |
0.0511795452 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 10 |
Hb_185830_050 |
0.0528477749 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 11 |
Hb_000334_290 |
0.0533332807 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 12 |
Hb_004525_010 |
0.0571370234 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 13 |
Hb_007559_020 |
0.0581229397 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 14 |
Hb_000083_020 |
0.0630942023 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 15 |
Hb_027403_010 |
0.0659327544 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 16 |
Hb_005605_020 |
0.0670518592 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 17 |
Hb_001046_060 |
0.0718753245 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_005605_010 |
0.0726130109 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 19 |
Hb_001104_080 |
0.0736862865 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 20 |
Hb_000825_010 |
0.0742372645 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |