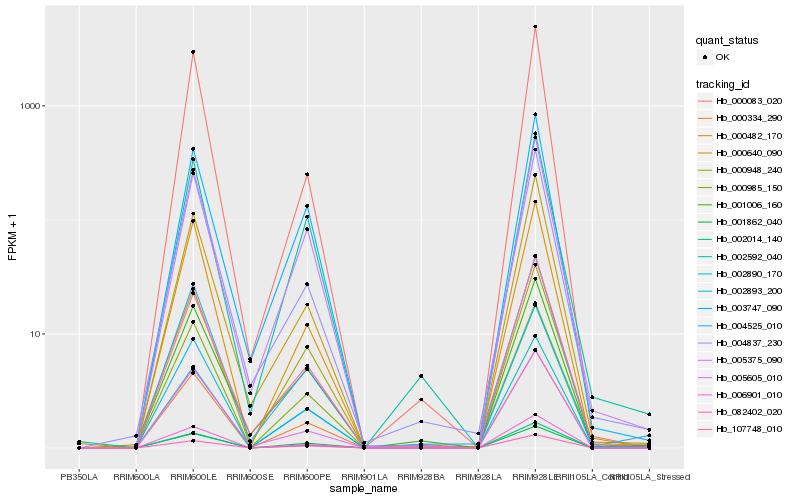
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_290 |
0.0 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 2 |
Hb_000985_150 |
0.0422546247 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 3 |
Hb_002014_140 |
0.0509303781 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_006901_010 |
0.052676518 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 5 |
Hb_002893_200 |
0.0533332807 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_004525_010 |
0.0569858337 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 7 |
Hb_082402_020 |
0.0574136544 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 8 |
Hb_000948_240 |
0.0582401628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000482_170 |
0.0632939266 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 10 |
Hb_002890_170 |
0.0640892535 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 11 |
Hb_107748_010 |
0.0673262821 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_005375_090 |
0.069216933 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001862_040 |
0.0703537095 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_001006_160 |
0.0749112565 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_003747_090 |
0.0750849531 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_005605_010 |
0.0764119287 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 17 |
Hb_002592_040 |
0.0773807843 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 18 |
Hb_000640_090 |
0.0785834377 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000083_020 |
0.0803747563 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 20 |
Hb_004837_230 |
0.08064277 |
- |
- |
unknown [Populus trichocarpa] |