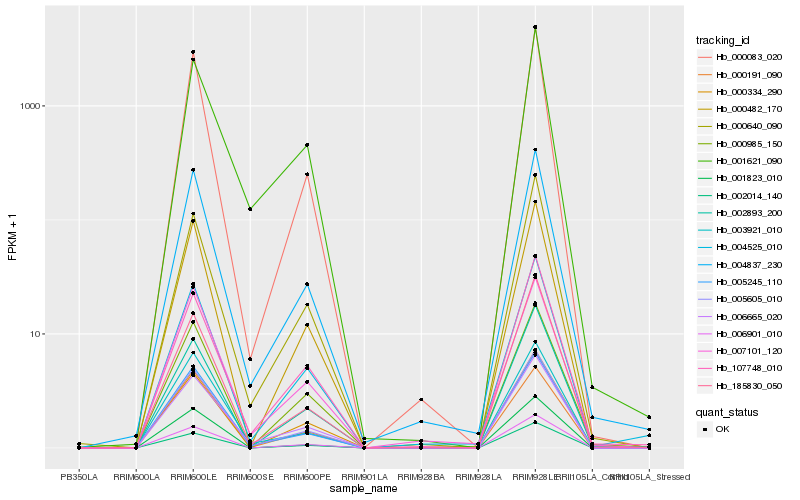
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005605_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 2 |
Hb_005245_110 |
0.030101833 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 3 |
Hb_000083_020 |
0.0365954722 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 4 |
Hb_000482_170 |
0.0411651244 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 5 |
Hb_004837_230 |
0.0451293799 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_006901_010 |
0.0486045207 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 7 |
Hb_003921_010 |
0.0568755257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006665_020 |
0.0589383913 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 9 |
Hb_002014_140 |
0.0602459305 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_001823_010 |
0.0627282146 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 11 |
Hb_004525_010 |
0.0660343669 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 12 |
Hb_000640_090 |
0.0700461432 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_002893_200 |
0.0726130109 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000985_150 |
0.0745556214 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 15 |
Hb_000334_290 |
0.0764119287 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 16 |
Hb_007101_120 |
0.0770280024 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 17 |
Hb_001621_090 |
0.078259989 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 18 |
Hb_000191_090 |
0.0785157745 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 19 |
Hb_185830_050 |
0.0814787368 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_107748_010 |
0.0827380338 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |