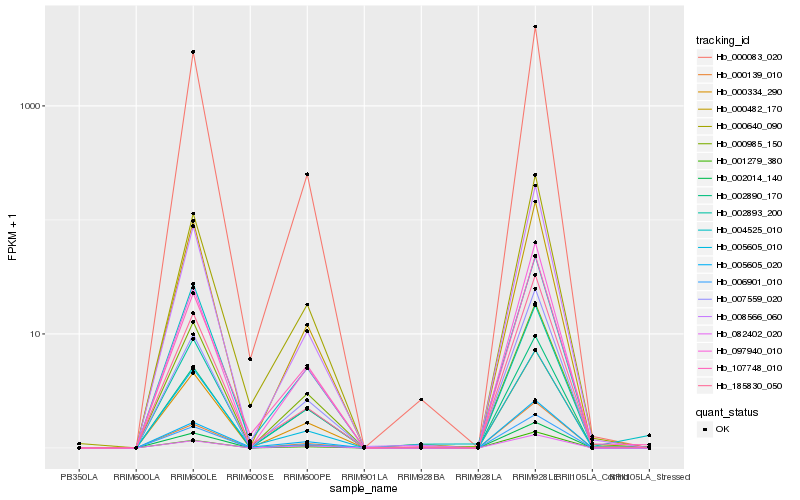
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002014_140 |
0.0 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_006901_010 |
0.0176389756 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 3 |
Hb_002893_200 |
0.038487835 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_001279_380 |
0.0411322099 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 5 |
Hb_000334_290 |
0.0509303781 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 6 |
Hb_000139_010 |
0.052780936 |
- |
- |
- |
| 7 |
Hb_000482_170 |
0.053228059 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 8 |
Hb_000083_020 |
0.054343853 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_000640_090 |
0.0552563561 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_097940_010 |
0.0557433276 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 11 |
Hb_008566_060 |
0.0571496429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005605_010 |
0.0602459305 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 13 |
Hb_082402_020 |
0.0606314136 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 14 |
Hb_002890_170 |
0.0633545239 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 15 |
Hb_007559_020 |
0.0639457811 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 16 |
Hb_107748_010 |
0.0649183472 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_004525_010 |
0.0660088248 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 18 |
Hb_185830_050 |
0.0697367649 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 19 |
Hb_000985_150 |
0.0718713404 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 20 |
Hb_005605_020 |
0.0723473272 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |