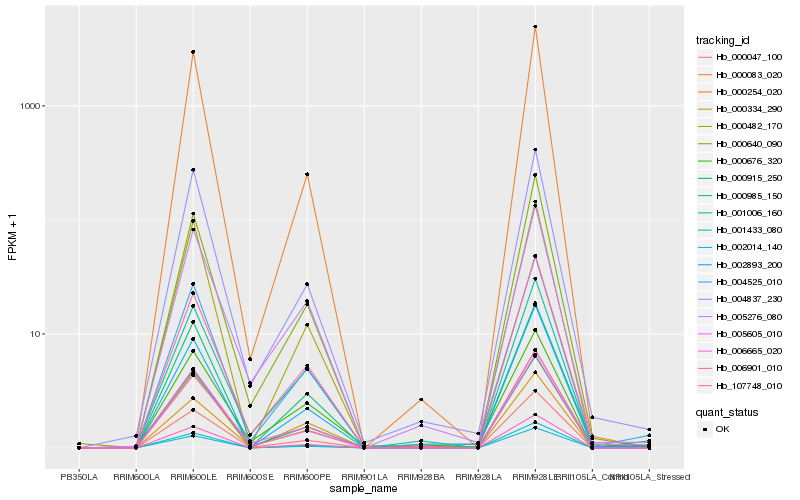
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_010 |
0.0 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 2 |
Hb_000334_290 |
0.0569858337 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 3 |
Hb_002893_200 |
0.0571370234 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_004837_230 |
0.0593466347 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_000985_150 |
0.0615641531 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 6 |
Hb_107748_010 |
0.0615966274 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_006901_010 |
0.064919192 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 8 |
Hb_002014_140 |
0.0660088248 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 9 |
Hb_005605_010 |
0.0660343669 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 10 |
Hb_001006_160 |
0.0674694952 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000640_090 |
0.0705222594 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_006665_020 |
0.0716365027 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 13 |
Hb_000083_020 |
0.0719322237 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_000482_170 |
0.0726823972 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 15 |
Hb_000254_020 |
0.0749378205 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 16 |
Hb_000047_100 |
0.0804800755 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005276_080 |
0.0810757911 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001433_080 |
0.0837117758 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 19 |
Hb_000676_320 |
0.0845805032 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 20 |
Hb_000915_250 |
0.0849784381 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |