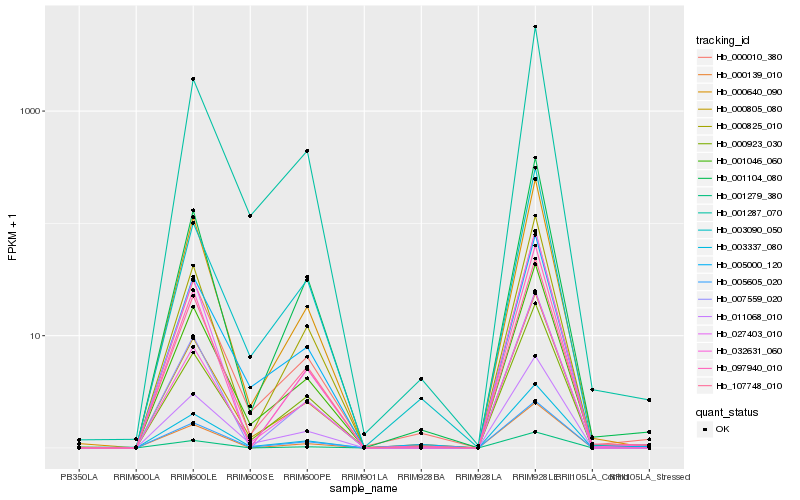
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_080 |
0.0 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000010_380 |
0.0342551743 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 3 |
Hb_001046_060 |
0.0365968146 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 4 |
Hb_003337_080 |
0.03711088 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 5 |
Hb_011068_010 |
0.0392479634 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 6 |
Hb_005605_020 |
0.0434681574 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 7 |
Hb_001287_070 |
0.0453143395 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 8 |
Hb_001104_080 |
0.0497420389 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 9 |
Hb_000923_030 |
0.0509089575 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 10 |
Hb_032631_060 |
0.0524251532 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_107748_010 |
0.0534339703 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000640_090 |
0.0567440334 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000825_010 |
0.0595318182 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 14 |
Hb_005000_120 |
0.0642718358 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 15 |
Hb_003090_050 |
0.0679757508 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 16 |
Hb_007559_020 |
0.0681114664 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 17 |
Hb_097940_010 |
0.0689367862 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 18 |
Hb_000139_010 |
0.0707832862 |
- |
- |
- |
| 19 |
Hb_027403_010 |
0.0725715598 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 20 |
Hb_001279_380 |
0.0741900106 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |