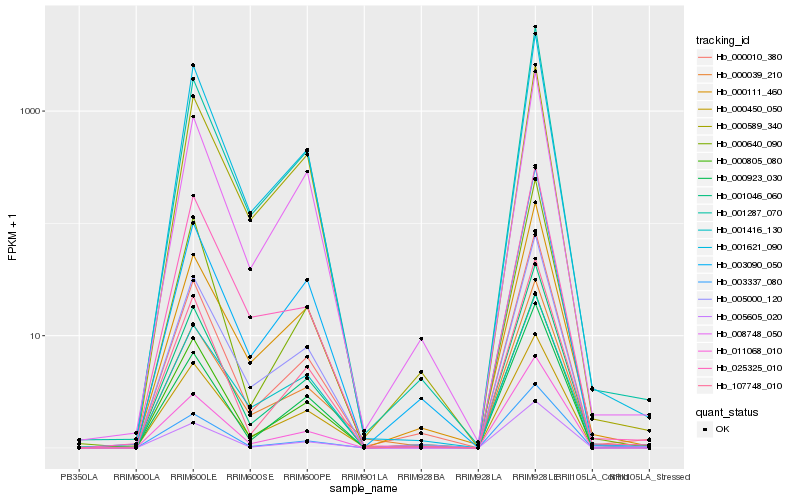
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_120 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 2 |
Hb_001046_060 |
0.0423682852 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001287_070 |
0.0435359668 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 4 |
Hb_001621_090 |
0.048746865 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 5 |
Hb_011068_010 |
0.0499835959 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 6 |
Hb_000039_210 |
0.0539339661 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 7 |
Hb_000111_460 |
0.0562681637 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 8 |
Hb_005605_020 |
0.0571928587 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 9 |
Hb_000450_050 |
0.0582607916 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 10 |
Hb_008748_050 |
0.0583033135 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 11 |
Hb_000805_080 |
0.0642718358 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000010_380 |
0.0655662908 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 13 |
Hb_003090_050 |
0.0666207279 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 14 |
Hb_003337_080 |
0.0680499902 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 15 |
Hb_107748_010 |
0.0694992713 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000589_340 |
0.0709345799 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 17 |
Hb_000923_030 |
0.0751208795 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 18 |
Hb_025325_010 |
0.0752707792 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 19 |
Hb_000640_090 |
0.0778873698 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001416_130 |
0.0792452052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |