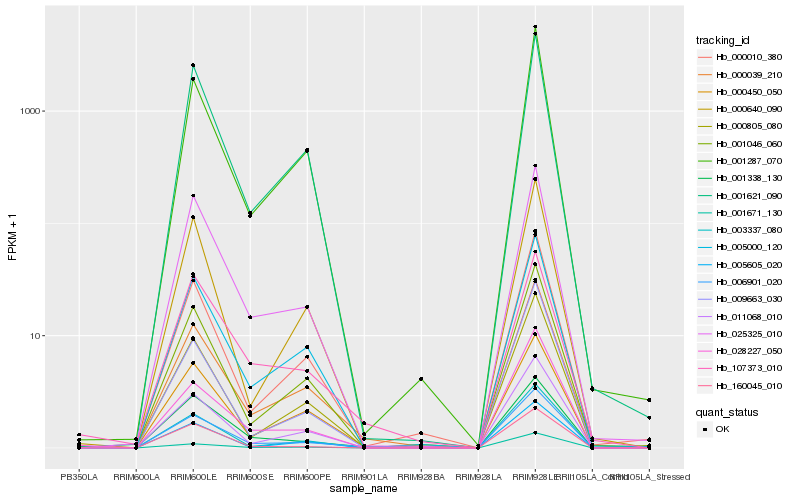
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006901_020 |
0.0 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 2 |
Hb_000039_210 |
0.0590010034 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 3 |
Hb_025325_010 |
0.0803650653 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 4 |
Hb_005000_120 |
0.0822929281 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 5 |
Hb_011068_010 |
0.0862358956 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 6 |
Hb_003337_080 |
0.0887917617 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 7 |
Hb_001287_070 |
0.0888182253 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 8 |
Hb_001046_060 |
0.091902894 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 9 |
Hb_001621_090 |
0.0958677568 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 10 |
Hb_000805_080 |
0.0959161272 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000010_380 |
0.0985780695 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 12 |
Hb_001338_130 |
0.0986925302 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 13 |
Hb_160045_010 |
0.0994034504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005605_020 |
0.1001320586 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 15 |
Hb_028227_050 |
0.10392587 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_000450_050 |
0.1040151236 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 17 |
Hb_001671_130 |
0.1058952645 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_000640_090 |
0.1105329961 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_009663_030 |
0.1112923199 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 20 |
Hb_107373_010 |
0.1127917505 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |