| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
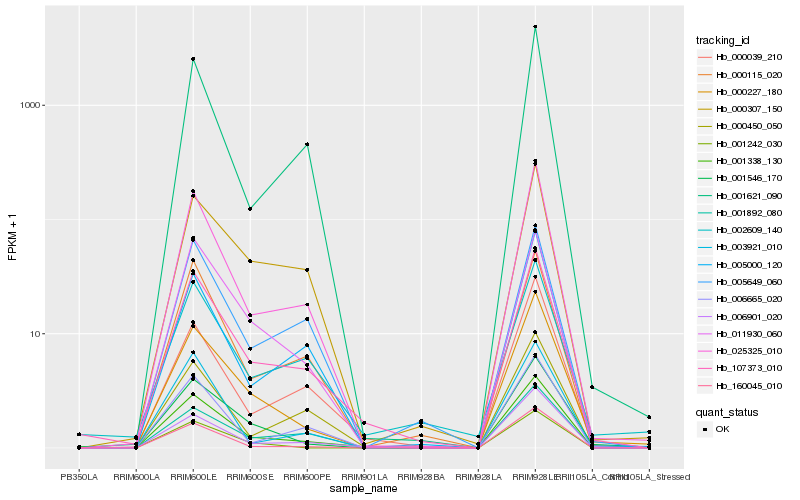
Hb_001338_130 |
0.0 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 2 |
Hb_107373_010 |
0.0618821631 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_025325_010 |
0.0844776382 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 4 |
Hb_006901_020 |
0.0986925302 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 5 |
Hb_000227_180 |
0.1061498552 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_160045_010 |
0.1106458355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011930_060 |
0.1118895128 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_000115_020 |
0.121106766 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_001621_090 |
0.122143313 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 10 |
Hb_001546_170 |
0.1228441462 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000307_150 |
0.1319981976 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001892_080 |
0.1321848434 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 13 |
Hb_006665_020 |
0.1322210402 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 14 |
Hb_000039_210 |
0.1322487281 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 15 |
Hb_001242_030 |
0.1322882249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005000_120 |
0.132857453 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 17 |
Hb_003921_010 |
0.1356126509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005649_060 |
0.1358954102 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 19 |
Hb_000450_050 |
0.1360719089 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 20 |
Hb_002609_140 |
0.1372694231 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |