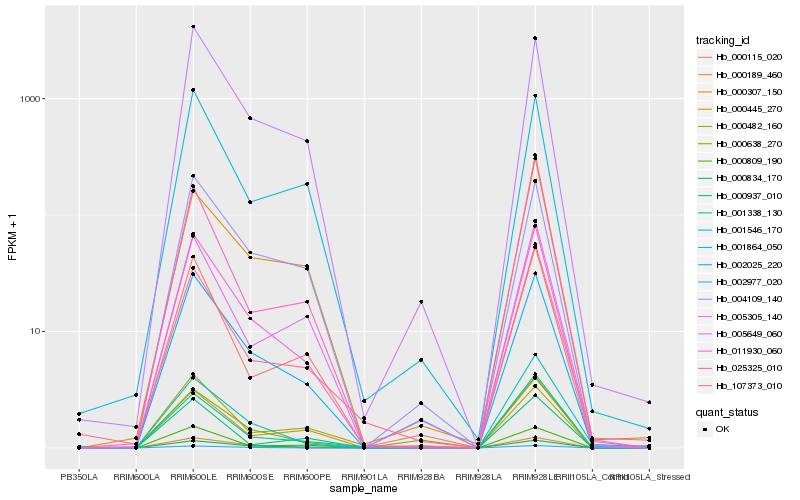
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011930_060 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_000834_170 |
0.076559879 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 3 |
Hb_002025_220 |
0.0868244065 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_005305_140 |
0.1006477579 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_000115_020 |
0.1082672013 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_001338_130 |
0.1118895128 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 7 |
Hb_107373_010 |
0.1135378066 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_004109_140 |
0.114090836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001864_050 |
0.1189890872 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 10 |
Hb_000482_160 |
0.1202974407 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 11 |
Hb_001546_170 |
0.1231964774 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_005649_060 |
0.1241210828 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 13 |
Hb_000638_270 |
0.1263976136 |
- |
- |
- |
| 14 |
Hb_002977_020 |
0.12674978 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 15 |
Hb_000307_150 |
0.1271869216 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000809_190 |
0.1276164917 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 17 |
Hb_000445_270 |
0.1282460486 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000189_460 |
0.1330311023 |
- |
- |
- |
| 19 |
Hb_025325_010 |
0.1331155496 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 20 |
Hb_000937_010 |
0.1334722126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |