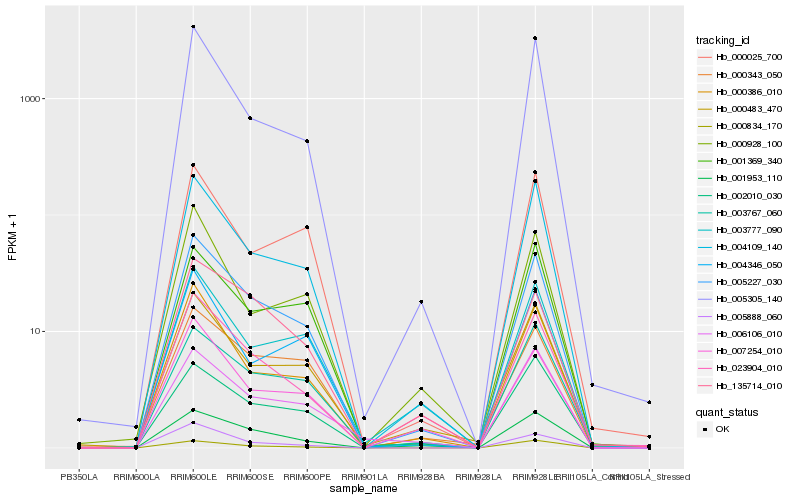
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005227_030 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004109_140 |
0.0737415731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000386_010 |
0.0876128685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005305_140 |
0.0929236976 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_005888_060 |
0.0956451425 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 6 |
Hb_001953_110 |
0.0960253331 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 7 |
Hb_023904_010 |
0.0972423406 |
- |
- |
- |
| 8 |
Hb_000483_470 |
0.0987886936 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000343_050 |
0.1050351725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007254_010 |
0.1056664781 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 11 |
Hb_003767_060 |
0.1092800855 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 12 |
Hb_000834_170 |
0.1094814375 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 13 |
Hb_003777_090 |
0.1125245514 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 14 |
Hb_006106_010 |
0.1184086568 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000025_700 |
0.1196573837 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 16 |
Hb_001369_340 |
0.1197614565 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 17 |
Hb_135714_010 |
0.1199749398 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 18 |
Hb_004346_050 |
0.1206127688 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 19 |
Hb_002010_030 |
0.1223125281 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_000928_100 |
0.1234308452 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |