| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
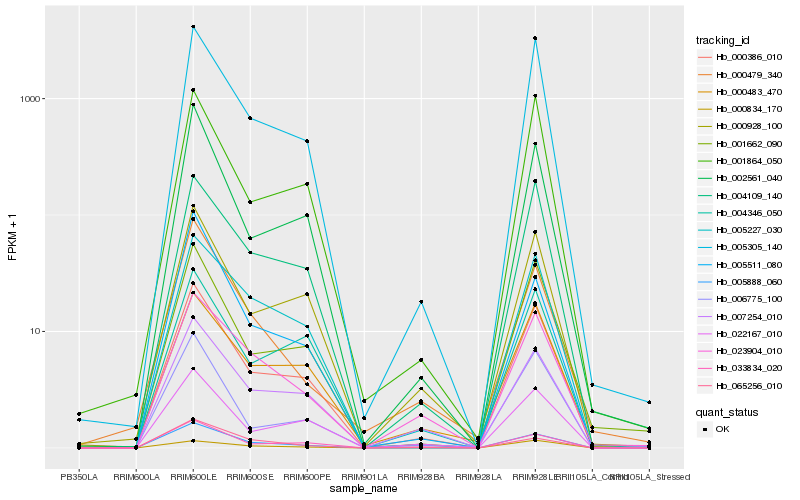
Hb_005888_060 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 2 |
Hb_000386_010 |
0.0848902976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005227_030 |
0.0956451425 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005305_140 |
0.1030027111 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_002561_040 |
0.1136732706 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 6 |
Hb_007254_010 |
0.1222370024 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 7 |
Hb_005511_080 |
0.1241418597 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 8 |
Hb_004109_140 |
0.1267560263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_033834_020 |
0.1287992243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_023904_010 |
0.1288548161 |
- |
- |
- |
| 11 |
Hb_000928_100 |
0.129236799 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 12 |
Hb_001662_090 |
0.1310280909 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_065256_010 |
0.1317101275 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 14 |
Hb_000479_340 |
0.1330249551 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 15 |
Hb_004346_050 |
0.1421507243 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 16 |
Hb_022167_010 |
0.1441736245 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 17 |
Hb_000834_170 |
0.147216837 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 18 |
Hb_000483_470 |
0.1508733582 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_006775_100 |
0.1521946266 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001864_050 |
0.1532530549 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |