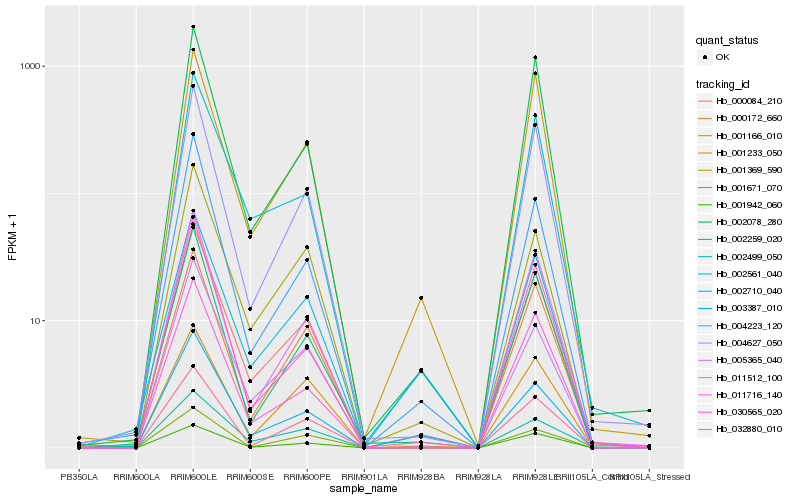
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_210 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 2 |
Hb_002561_040 |
0.0712990684 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 3 |
Hb_002499_050 |
0.0734149795 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_004627_050 |
0.0758941355 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 5 |
Hb_001369_590 |
0.0806019609 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 6 |
Hb_002259_020 |
0.0831402805 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 7 |
Hb_011512_100 |
0.0834081139 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005365_040 |
0.0843744618 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 9 |
Hb_004223_120 |
0.0857641401 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000172_660 |
0.0883356538 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_001942_060 |
0.0907477746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002078_280 |
0.0949809475 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 13 |
Hb_001233_050 |
0.0957432461 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_002710_040 |
0.0963991545 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 15 |
Hb_011716_140 |
0.0974482379 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 16 |
Hb_030565_020 |
0.0977244374 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 17 |
Hb_003387_010 |
0.0997614226 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 18 |
Hb_032880_010 |
0.1002828584 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 19 |
Hb_001166_010 |
0.1003493408 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 20 |
Hb_001671_070 |
0.1015158953 |
- |
- |
kinase, putative [Ricinus communis] |