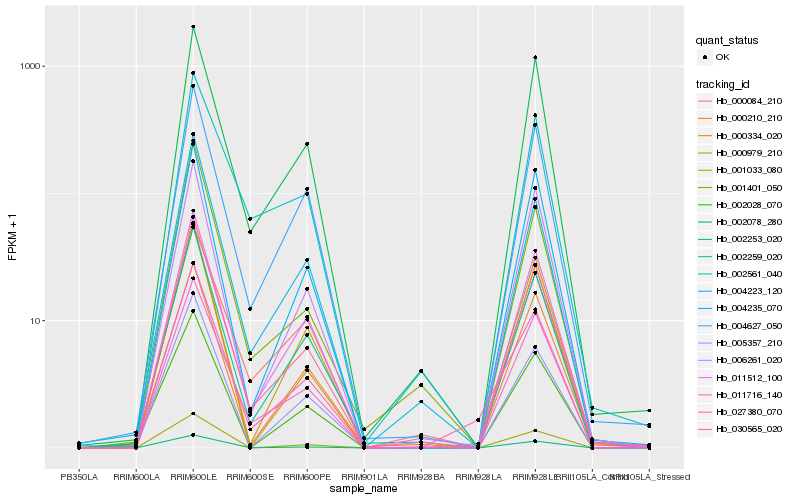
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030565_020 |
0.0 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 2 |
Hb_011716_140 |
0.0568360962 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 3 |
Hb_002253_020 |
0.0573051307 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 4 |
Hb_011512_100 |
0.0612761495 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002259_020 |
0.0630163703 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 6 |
Hb_004223_120 |
0.0699570032 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002078_280 |
0.0722008518 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 8 |
Hb_004627_050 |
0.0761636048 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 9 |
Hb_001033_080 |
0.0821248537 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 10 |
Hb_001401_050 |
0.0857096325 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006261_020 |
0.0903402462 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 12 |
Hb_000334_020 |
0.0913266272 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_005357_210 |
0.0935160241 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 14 |
Hb_002561_040 |
0.0951693638 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 15 |
Hb_000979_210 |
0.0956228386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000210_210 |
0.0961596253 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004235_070 |
0.0963132501 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 18 |
Hb_027380_070 |
0.0963880937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000084_210 |
0.0977244374 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 20 |
Hb_002028_070 |
0.0978747574 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |