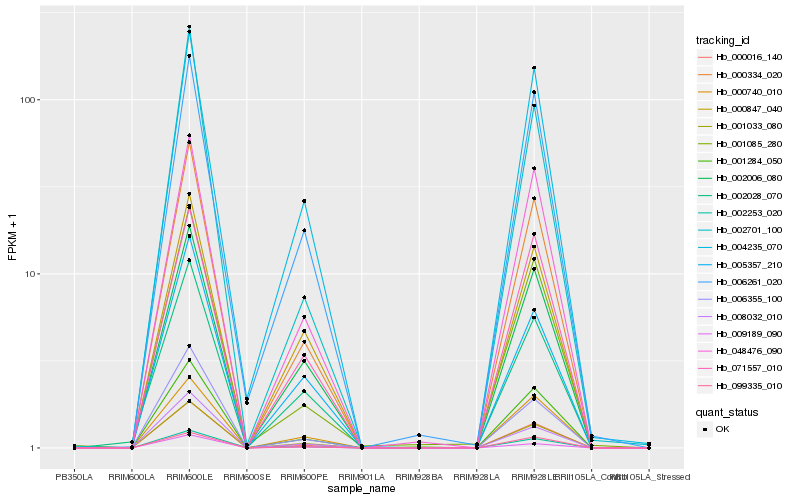
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_020 |
0.0 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 2 |
Hb_000016_140 |
0.0215394892 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_001033_080 |
0.0280822748 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 4 |
Hb_001284_050 |
0.0536754738 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 5 |
Hb_002253_020 |
0.0610772213 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 6 |
Hb_048476_090 |
0.0683168789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001085_280 |
0.0684825286 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 8 |
Hb_006355_100 |
0.0717496353 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 9 |
Hb_002006_080 |
0.0748863114 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 10 |
Hb_002701_100 |
0.0755112332 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 11 |
Hb_099335_010 |
0.0767331802 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 12 |
Hb_000740_010 |
0.0785155827 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_002028_070 |
0.0801498062 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 14 |
Hb_006261_020 |
0.0806807175 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 15 |
Hb_004235_070 |
0.0819983187 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 16 |
Hb_009189_090 |
0.0823378661 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 17 |
Hb_008032_010 |
0.0838771107 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 18 |
Hb_000847_040 |
0.084411681 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 19 |
Hb_005357_210 |
0.0900160694 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 20 |
Hb_071557_010 |
0.0908047414 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |