| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
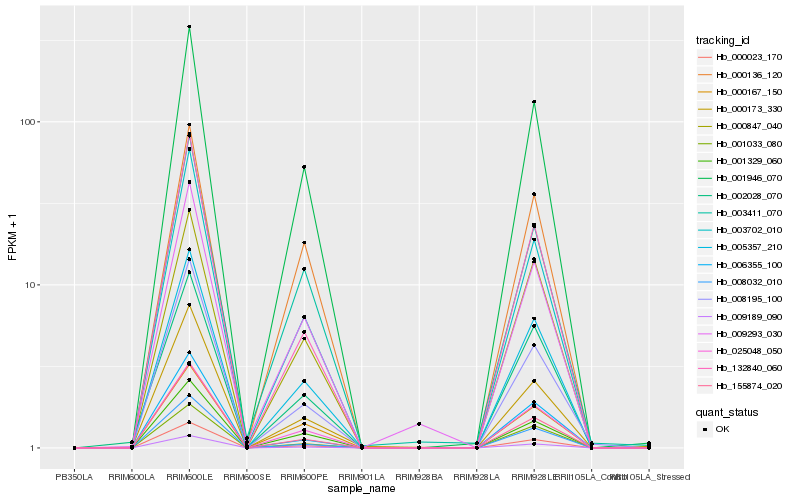
Hb_005357_210 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 2 |
Hb_025048_050 |
0.0253812554 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 3 |
Hb_001946_070 |
0.0374015851 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000023_170 |
0.0434380581 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 5 |
Hb_009189_090 |
0.0454492921 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 6 |
Hb_003702_010 |
0.0472429453 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 7 |
Hb_001329_060 |
0.0515390233 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_001033_080 |
0.0630712365 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 9 |
Hb_002028_070 |
0.0655197145 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 10 |
Hb_000847_040 |
0.0687762672 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 11 |
Hb_000136_120 |
0.0701956156 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 12 |
Hb_000167_150 |
0.0713693549 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 13 |
Hb_008032_010 |
0.0716231214 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 14 |
Hb_003411_070 |
0.073328428 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 15 |
Hb_009293_030 |
0.0739559615 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 16 |
Hb_132840_060 |
0.0744660311 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 17 |
Hb_000173_330 |
0.077035228 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 18 |
Hb_006355_100 |
0.0789171022 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 19 |
Hb_008195_100 |
0.0790657549 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 20 |
Hb_155874_020 |
0.0800677369 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |