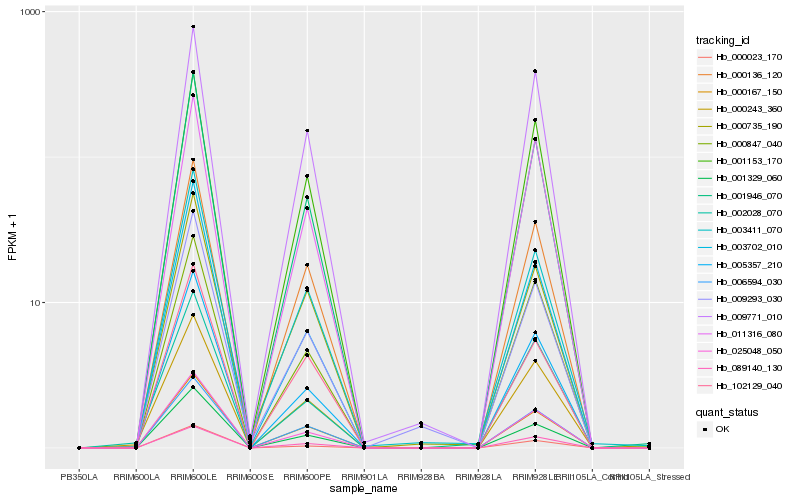
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_070 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_025048_050 |
0.0180767976 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 3 |
Hb_001329_060 |
0.0347129896 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 4 |
Hb_005357_210 |
0.0374015851 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 5 |
Hb_000167_150 |
0.0393561575 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 6 |
Hb_000136_120 |
0.0395094967 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 7 |
Hb_006594_030 |
0.0553539595 |
- |
- |
- |
| 8 |
Hb_003411_070 |
0.0587441477 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 9 |
Hb_089140_130 |
0.0626448651 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 10 |
Hb_000847_040 |
0.0634093272 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 11 |
Hb_001153_170 |
0.0645308013 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 12 |
Hb_009293_030 |
0.0676585273 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 13 |
Hb_003702_010 |
0.0686230033 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 14 |
Hb_000243_360 |
0.0687836008 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 15 |
Hb_011316_080 |
0.0698378683 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 16 |
Hb_000023_170 |
0.0720140483 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 17 |
Hb_000735_190 |
0.0725039269 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 18 |
Hb_002028_070 |
0.073701305 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 19 |
Hb_102129_040 |
0.0742801874 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009771_010 |
0.0747881969 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |