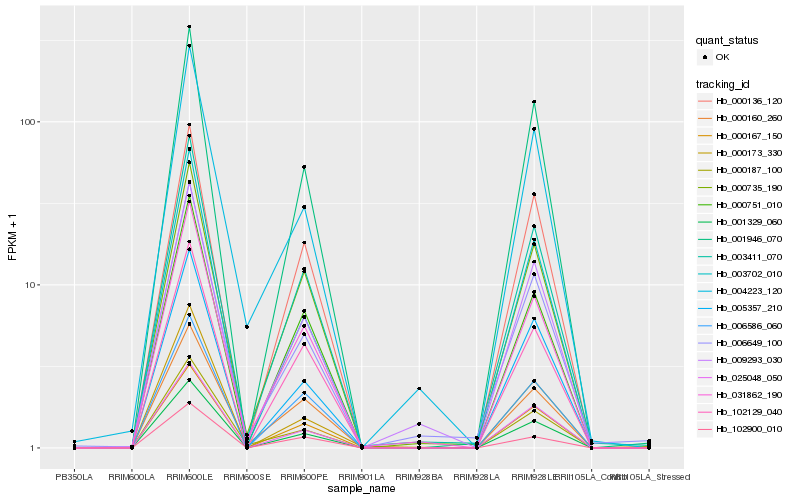
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003411_070 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 2 |
Hb_031862_190 |
0.0413268443 |
- |
- |
- |
| 3 |
Hb_001329_060 |
0.0457849585 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 4 |
Hb_000751_010 |
0.0532694666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000735_190 |
0.054488392 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 6 |
Hb_001946_070 |
0.0587441477 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_009293_030 |
0.0608107419 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 8 |
Hb_102129_040 |
0.0613779357 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006649_100 |
0.0655442774 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_000136_120 |
0.0687981113 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 11 |
Hb_000167_150 |
0.0691015563 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 12 |
Hb_025048_050 |
0.0691291869 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 13 |
Hb_006586_060 |
0.0707006991 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 14 |
Hb_005357_210 |
0.073328428 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 15 |
Hb_000187_100 |
0.0747846008 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_000160_260 |
0.07536426 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 17 |
Hb_003702_010 |
0.0756299564 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 18 |
Hb_102900_010 |
0.0825623631 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000173_330 |
0.0832369446 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 20 |
Hb_004223_120 |
0.0845060809 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |