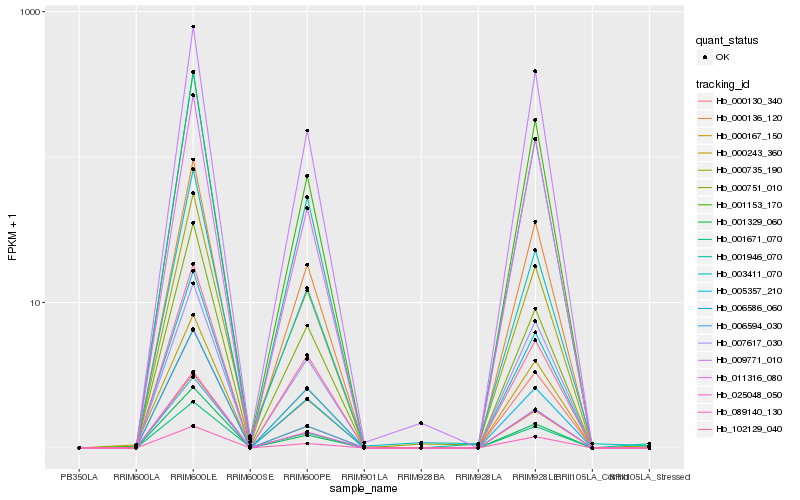
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000167_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 2 |
Hb_000136_120 |
0.0142584506 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 3 |
Hb_006594_030 |
0.0250310854 |
- |
- |
- |
| 4 |
Hb_001946_070 |
0.0393561575 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001329_060 |
0.0460651874 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 6 |
Hb_006586_060 |
0.0478523899 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_025048_050 |
0.049520647 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 8 |
Hb_001153_170 |
0.0500353718 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 9 |
Hb_089140_130 |
0.0539366875 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 10 |
Hb_000735_190 |
0.0551961425 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 11 |
Hb_102129_040 |
0.0567012742 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009771_010 |
0.061929963 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 13 |
Hb_000130_340 |
0.0654314596 |
- |
- |
- |
| 14 |
Hb_011316_080 |
0.0682371256 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 15 |
Hb_003411_070 |
0.0691015563 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 16 |
Hb_000243_360 |
0.0706203167 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 17 |
Hb_000751_010 |
0.0707768008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005357_210 |
0.0713693549 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 19 |
Hb_001671_070 |
0.0723864763 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_007617_030 |
0.0734213965 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |