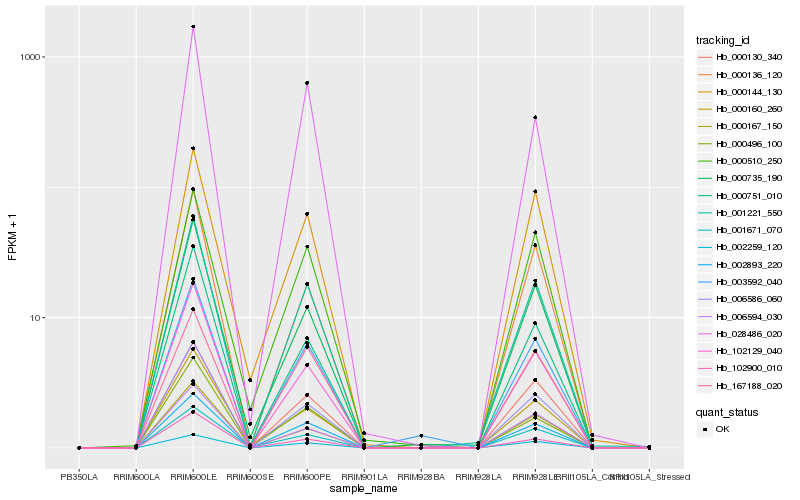
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_550 |
0.0 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002893_220 |
0.0435077324 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |
| 3 |
Hb_006586_060 |
0.0561598942 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000130_340 |
0.0637628598 |
- |
- |
- |
| 5 |
Hb_000735_190 |
0.0699654789 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 6 |
Hb_003592_040 |
0.070503305 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 7 |
Hb_102129_040 |
0.071569808 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002259_120 |
0.077052074 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 9 |
Hb_001671_070 |
0.0780174586 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000160_260 |
0.0792168382 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 11 |
Hb_000167_150 |
0.0847509317 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 12 |
Hb_000496_100 |
0.08727638 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 13 |
Hb_006594_030 |
0.0872881852 |
- |
- |
- |
| 14 |
Hb_000751_010 |
0.087526221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000136_120 |
0.088005886 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 16 |
Hb_028486_020 |
0.0904917941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000144_130 |
0.0924342082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_167188_020 |
0.0932742827 |
- |
- |
PREDICTED: uncharacterized protein At1g05835 [Jatropha curcas] |
| 19 |
Hb_000510_250 |
0.0936243415 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 20 |
Hb_102900_010 |
0.0946149869 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |