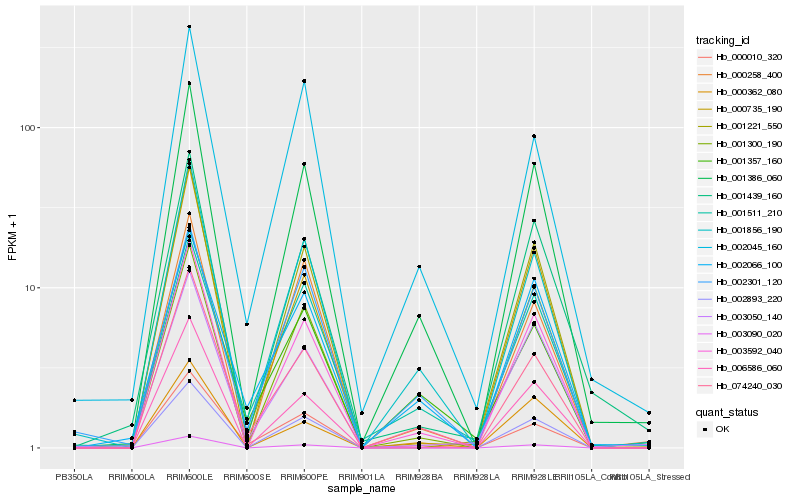
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001386_060 |
0.0 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 2 |
Hb_001856_190 |
0.0572948954 |
- |
- |
SP1L, putative [Ricinus communis] |
| 3 |
Hb_003592_040 |
0.0634050554 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 4 |
Hb_003090_020 |
0.0748717836 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 5 |
Hb_074240_030 |
0.0820652668 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 6 |
Hb_001439_160 |
0.1018276902 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 7 |
Hb_001221_550 |
0.1049029115 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_001357_160 |
0.1068672397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000010_320 |
0.1082004093 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 10 |
Hb_002893_220 |
0.1093382149 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |
| 11 |
Hb_000258_400 |
0.1142279105 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 12 |
Hb_001511_210 |
0.1144603225 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 13 |
Hb_002066_100 |
0.1149401126 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 14 |
Hb_003050_140 |
0.1149701885 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 15 |
Hb_002045_160 |
0.1169046182 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 16 |
Hb_000735_190 |
0.1171355674 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 17 |
Hb_000362_080 |
0.1176461722 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 18 |
Hb_001300_190 |
0.1178829306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002301_120 |
0.1185648364 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 20 |
Hb_006586_060 |
0.1216197812 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |