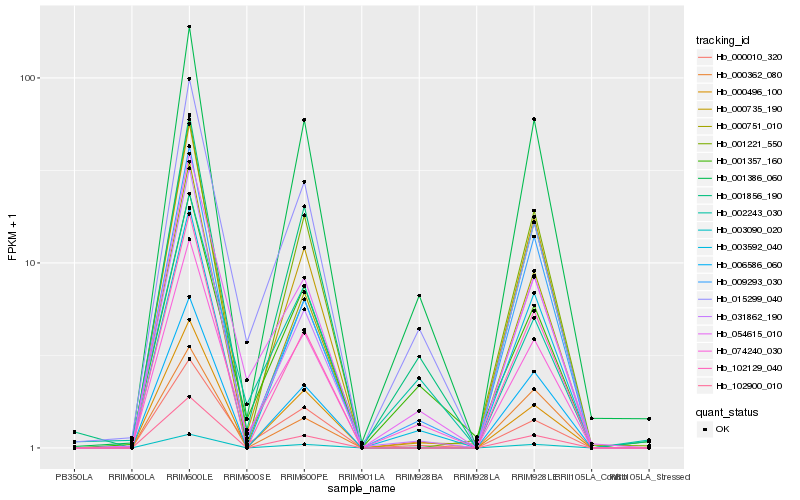
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 2 |
Hb_001856_190 |
0.0686888516 |
- |
- |
SP1L, putative [Ricinus communis] |
| 3 |
Hb_001386_060 |
0.0748717836 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 4 |
Hb_074240_030 |
0.0791809079 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 5 |
Hb_003592_040 |
0.0818936213 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 6 |
Hb_001357_160 |
0.1047045296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000362_080 |
0.1154958927 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 8 |
Hb_000010_320 |
0.1215291343 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 9 |
Hb_015299_040 |
0.1223376407 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 10 |
Hb_006586_060 |
0.1290613125 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 11 |
Hb_102129_040 |
0.1303889038 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000735_190 |
0.13097669 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 13 |
Hb_009293_030 |
0.13228749 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 14 |
Hb_001221_550 |
0.1337146072 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_102900_010 |
0.133916297 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 16 |
Hb_031862_190 |
0.1339725477 |
- |
- |
- |
| 17 |
Hb_000496_100 |
0.1345182692 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 18 |
Hb_000751_010 |
0.1352959551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_054615_010 |
0.1359995525 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 20 |
Hb_002243_030 |
0.1362605248 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |