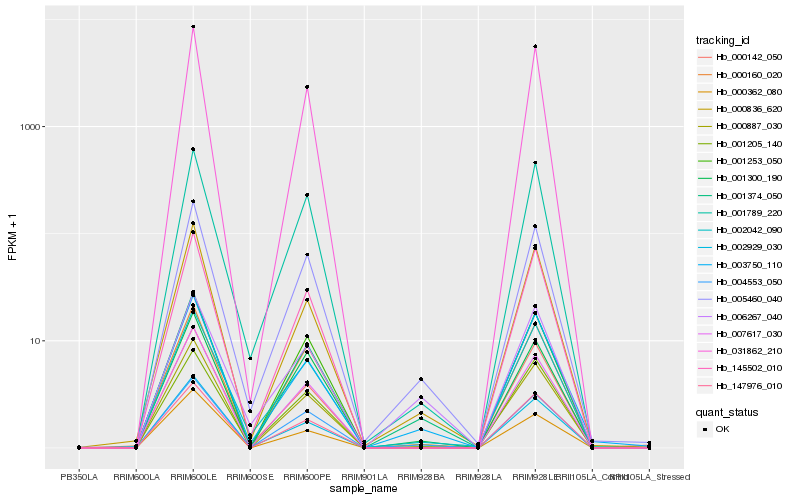
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001374_050 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 2 |
Hb_004553_050 |
0.0347123673 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 3 |
Hb_005460_040 |
0.0530182411 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 4 |
Hb_003750_110 |
0.0754700953 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 5 |
Hb_001300_190 |
0.0790333427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000836_620 |
0.0866099781 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001205_140 |
0.0892734655 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 8 |
Hb_000362_080 |
0.0894564929 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 9 |
Hb_002042_090 |
0.0913182774 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 10 |
Hb_031862_210 |
0.0932588215 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 11 |
Hb_006267_040 |
0.0932813014 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001789_220 |
0.0956308576 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 13 |
Hb_002929_030 |
0.0959955596 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 14 |
Hb_001253_050 |
0.0966819653 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 15 |
Hb_145502_010 |
0.0969096762 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 16 |
Hb_007617_030 |
0.0992423383 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 17 |
Hb_000887_030 |
0.0994538936 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_147976_010 |
0.0994755641 |
- |
- |
- |
| 19 |
Hb_000160_020 |
0.1023218178 |
- |
- |
PREDICTED: gamma-interferon-inducible lysosomal thiol reductase-like [Jatropha curcas] |
| 20 |
Hb_000142_050 |
0.1033386599 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |