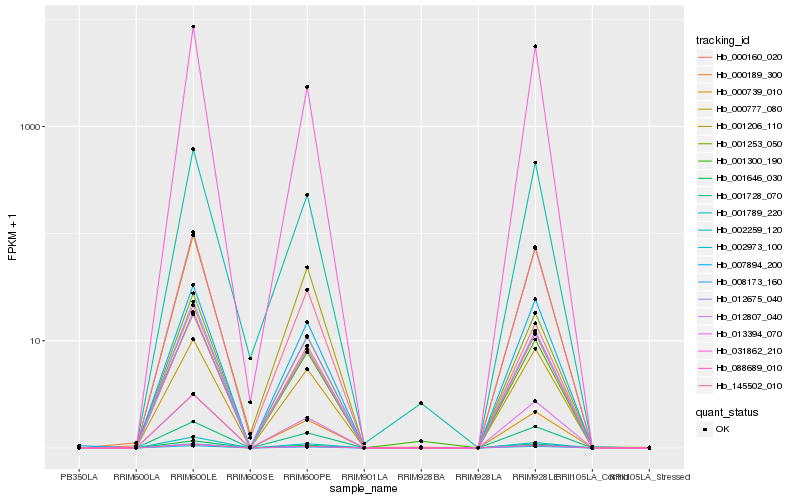
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001253_050 |
0.0 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 2 |
Hb_002973_100 |
0.0278884262 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Populus euphratica] |
| 3 |
Hb_008173_160 |
0.0292981992 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 4 |
Hb_088689_010 |
0.0293497997 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 5 |
Hb_012807_040 |
0.0302265738 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 6 |
Hb_000160_020 |
0.0308651483 |
- |
- |
PREDICTED: gamma-interferon-inducible lysosomal thiol reductase-like [Jatropha curcas] |
| 7 |
Hb_007894_200 |
0.0389924874 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |
| 8 |
Hb_000739_010 |
0.0412410849 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 9 |
Hb_001646_030 |
0.0423611361 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 10 |
Hb_013394_070 |
0.0451791354 |
- |
- |
hypothetical protein POPTR_0013s12090g [Populus trichocarpa] |
| 11 |
Hb_000777_080 |
0.0509549544 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 12 |
Hb_001728_070 |
0.0530508997 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 13 |
Hb_031862_210 |
0.0532579481 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 14 |
Hb_001206_110 |
0.0549108186 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 15 |
Hb_000189_300 |
0.0552302393 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001789_220 |
0.0579233464 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 17 |
Hb_145502_010 |
0.0581616528 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 18 |
Hb_001300_190 |
0.0638845948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012675_040 |
0.0638933104 |
- |
- |
PREDICTED: uncharacterized protein LOC105647773 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002259_120 |
0.0641945301 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |