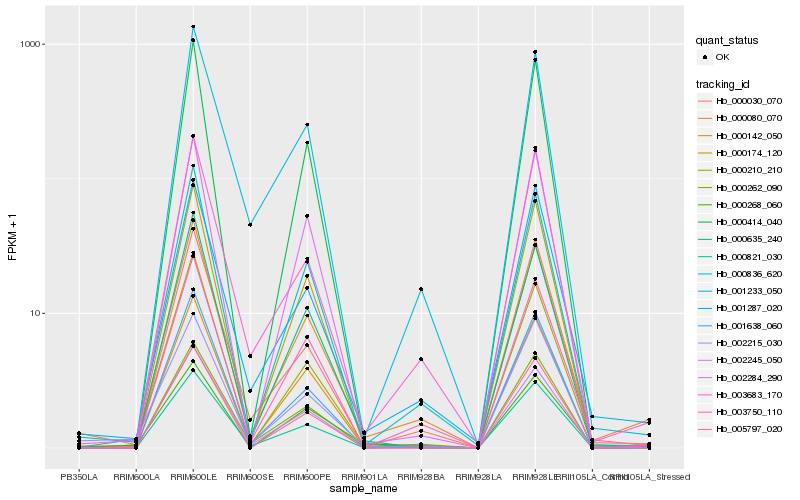
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_070 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003750_110 |
0.0782488348 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 3 |
Hb_001287_020 |
0.0785195389 |
- |
- |
SulA [Manihot esculenta] |
| 4 |
Hb_000836_620 |
0.0836579358 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001638_060 |
0.0862028404 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000142_050 |
0.0877070049 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 7 |
Hb_003683_170 |
0.0881157927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000030_070 |
0.0884030011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000210_210 |
0.0923683599 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000821_030 |
0.0942559236 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 11 |
Hb_000262_090 |
0.0979210449 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 12 |
Hb_000268_060 |
0.0995880858 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000174_120 |
0.1003615859 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 14 |
Hb_002215_030 |
0.1045180192 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001233_050 |
0.1048872672 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000414_040 |
0.1058820575 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 17 |
Hb_002245_050 |
0.1081616215 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_002284_290 |
0.1081902926 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 19 |
Hb_000635_240 |
0.1085659336 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 20 |
Hb_005797_020 |
0.1093054394 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |