| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
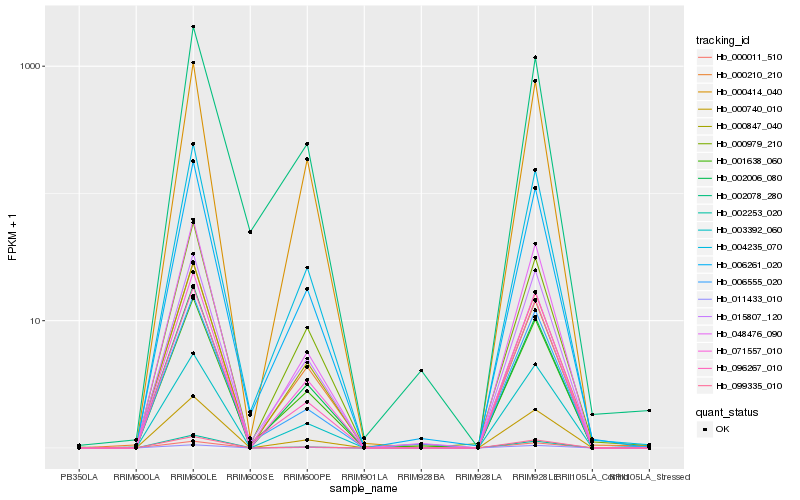
Hb_004235_070 |
0.0 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 2 |
Hb_006261_020 |
0.0237939753 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 3 |
Hb_000740_010 |
0.0374701337 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_015807_120 |
0.0388332494 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_071557_010 |
0.0427633043 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 6 |
Hb_001638_060 |
0.0445913607 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 7 |
Hb_002006_080 |
0.0504716048 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 8 |
Hb_048476_090 |
0.0514105094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000210_210 |
0.0534189408 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 10 |
Hb_099335_010 |
0.0546015099 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 11 |
Hb_000979_210 |
0.0587436381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003392_060 |
0.0595451713 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_096267_010 |
0.0640140631 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 14 |
Hb_002253_020 |
0.0647747816 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 15 |
Hb_000011_510 |
0.0661329705 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 16 |
Hb_002078_280 |
0.0676321998 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 17 |
Hb_000414_040 |
0.0677253584 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 18 |
Hb_000847_040 |
0.072407272 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 19 |
Hb_006555_020 |
0.0735632469 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 20 |
Hb_011433_010 |
0.0736639275 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |