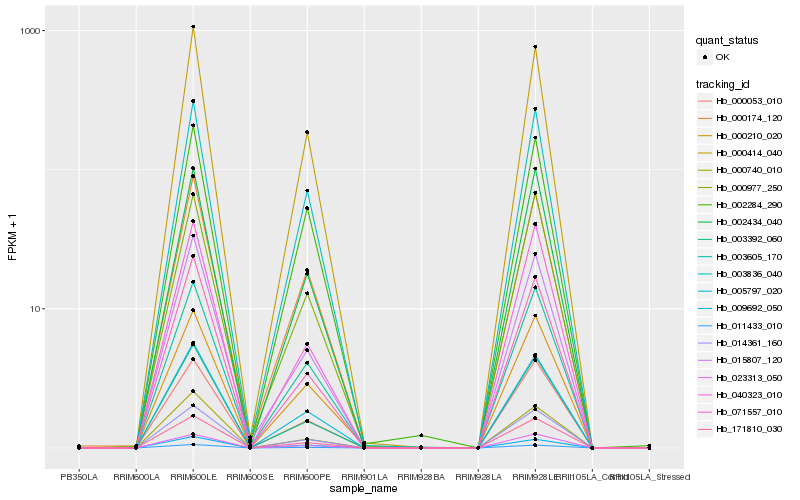
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011433_010 |
0.0 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 2 |
Hb_005797_020 |
0.0184058727 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 3 |
Hb_014361_160 |
0.0192634706 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 4 |
Hb_003392_060 |
0.0264612045 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 5 |
Hb_000414_040 |
0.0319011269 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 6 |
Hb_171810_030 |
0.0339547765 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 7 |
Hb_000053_010 |
0.0374178506 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 8 |
Hb_003605_170 |
0.0416975996 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 9 |
Hb_002434_040 |
0.04178654 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 10 |
Hb_015807_120 |
0.0426682524 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003836_040 |
0.0437662338 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 12 |
Hb_000174_120 |
0.0446975131 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_023313_050 |
0.0479166521 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 14 |
Hb_071557_010 |
0.0481727153 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 15 |
Hb_000210_020 |
0.0495714658 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 16 |
Hb_000977_250 |
0.0532870513 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 17 |
Hb_009692_050 |
0.0554841235 |
- |
- |
- |
| 18 |
Hb_040323_010 |
0.0560839197 |
- |
- |
- |
| 19 |
Hb_000740_010 |
0.0588623565 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_002284_290 |
0.0619364813 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |