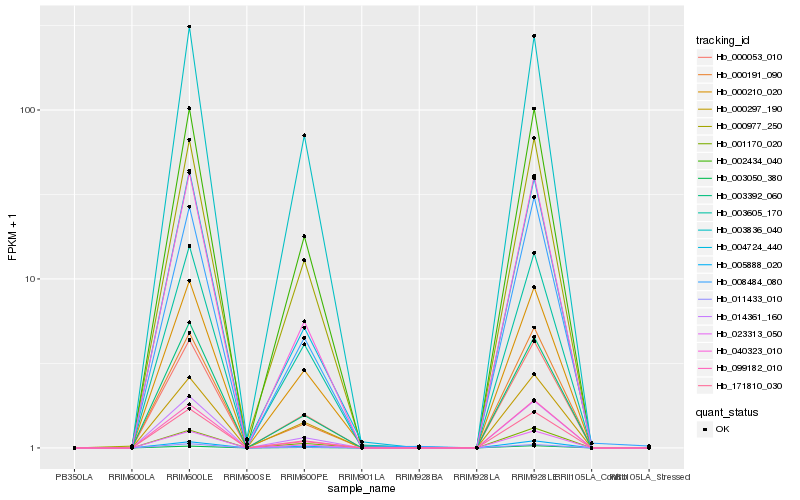
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023313_050 |
0.0 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 2 |
Hb_002434_040 |
0.0068234683 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 3 |
Hb_000053_010 |
0.0127812532 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 4 |
Hb_000977_250 |
0.0271452883 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 5 |
Hb_171810_030 |
0.0310317419 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 6 |
Hb_014361_160 |
0.03159643 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 7 |
Hb_099182_010 |
0.0381502178 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 8 |
Hb_040323_010 |
0.0389325162 |
- |
- |
- |
| 9 |
Hb_001170_020 |
0.0420484303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011433_010 |
0.0479166521 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 11 |
Hb_008484_080 |
0.0484779293 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_003605_170 |
0.0502206028 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 13 |
Hb_003050_380 |
0.0516488511 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 14 |
Hb_000297_190 |
0.0546058245 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 15 |
Hb_000210_020 |
0.0566265998 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 16 |
Hb_005888_020 |
0.0572016944 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 17 |
Hb_003392_060 |
0.0579399502 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_003836_040 |
0.0579994937 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 19 |
Hb_000191_090 |
0.0583280076 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 20 |
Hb_004724_440 |
0.0601842975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |